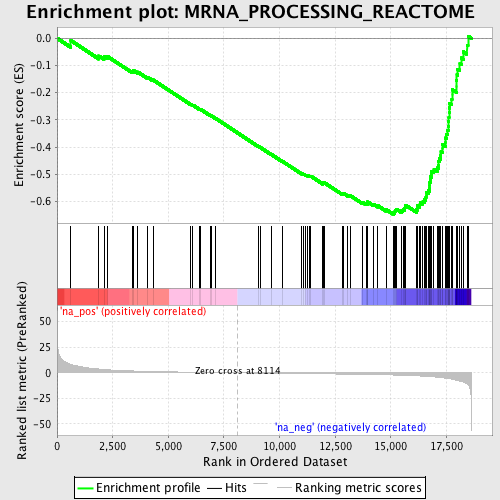
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MRNA_PROCESSING_REACTOME |
| Enrichment Score (ES) | -0.6475924 |
| Normalized Enrichment Score (NES) | -2.269057 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDIT3 | 618 | 8.219 | -0.0083 | No | ||
| 2 | CD2BP2 | 1878 | 3.694 | -0.0651 | No | ||
| 3 | HNRPC | 2108 | 3.252 | -0.0676 | No | ||
| 4 | CUGBP2 | 2256 | 3.022 | -0.0663 | No | ||
| 5 | RBM5 | 3387 | 1.824 | -0.1218 | No | ||
| 6 | COL2A1 | 3416 | 1.804 | -0.1178 | No | ||
| 7 | PTBP2 | 3624 | 1.670 | -0.1239 | No | ||
| 8 | SFRS7 | 4053 | 1.412 | -0.1427 | No | ||
| 9 | HNRPL | 4319 | 1.267 | -0.1531 | No | ||
| 10 | CSTF2 | 6000 | 0.620 | -0.2420 | No | ||
| 11 | PRPF4B | 6096 | 0.596 | -0.2453 | No | ||
| 12 | HNRPD | 6399 | 0.500 | -0.2601 | No | ||
| 13 | SF4 | 6456 | 0.483 | -0.2616 | No | ||
| 14 | CPSF2 | 6887 | 0.358 | -0.2837 | No | ||
| 15 | SF3B1 | 6934 | 0.342 | -0.2852 | No | ||
| 16 | SRPK2 | 7114 | 0.291 | -0.2940 | No | ||
| 17 | PABPN1 | 9064 | -0.243 | -0.3985 | No | ||
| 18 | SFRS14 | 9136 | -0.260 | -0.4015 | No | ||
| 19 | SNRPE | 9624 | -0.376 | -0.4267 | No | ||
| 20 | SUPT5H | 10128 | -0.510 | -0.4523 | No | ||
| 21 | CPSF3 | 10994 | -0.714 | -0.4968 | No | ||
| 22 | CLK4 | 11089 | -0.736 | -0.4996 | No | ||
| 23 | SNRPB2 | 11186 | -0.762 | -0.5025 | No | ||
| 24 | CSTF2T | 11260 | -0.777 | -0.5041 | No | ||
| 25 | SRRM1 | 11348 | -0.799 | -0.5064 | No | ||
| 26 | DNAJC8 | 11408 | -0.815 | -0.5071 | No | ||
| 27 | SF3B2 | 11949 | -0.956 | -0.5333 | No | ||
| 28 | NONO | 11953 | -0.957 | -0.5305 | No | ||
| 29 | SFRS16 | 12021 | -0.975 | -0.5312 | No | ||
| 30 | SNRPN | 12824 | -1.207 | -0.5708 | No | ||
| 31 | DHX8 | 12854 | -1.215 | -0.5687 | No | ||
| 32 | CLK2 | 13069 | -1.282 | -0.5764 | No | ||
| 33 | FUS | 13173 | -1.312 | -0.5779 | No | ||
| 34 | SF3B4 | 13725 | -1.497 | -0.6031 | No | ||
| 35 | HNRPAB | 13898 | -1.567 | -0.6076 | No | ||
| 36 | HNRPH2 | 13948 | -1.586 | -0.6054 | No | ||
| 37 | CSTF1 | 13963 | -1.592 | -0.6014 | No | ||
| 38 | HNRPR | 14218 | -1.684 | -0.6099 | No | ||
| 39 | SPOP | 14410 | -1.766 | -0.6149 | No | ||
| 40 | DHX15 | 14785 | -1.937 | -0.6292 | No | ||
| 41 | U2AF2 | 15127 | -2.117 | -0.6411 | Yes | ||
| 42 | SFRS9 | 15184 | -2.148 | -0.6376 | Yes | ||
| 43 | SFRS8 | 15199 | -2.156 | -0.6318 | Yes | ||
| 44 | NUDT21 | 15254 | -2.189 | -0.6281 | Yes | ||
| 45 | GIPC1 | 15480 | -2.336 | -0.6331 | Yes | ||
| 46 | PTBP1 | 15547 | -2.383 | -0.6294 | Yes | ||
| 47 | DICER1 | 15618 | -2.432 | -0.6258 | Yes | ||
| 48 | RNGTT | 15637 | -2.444 | -0.6193 | Yes | ||
| 49 | SNRPG | 15665 | -2.464 | -0.6133 | Yes | ||
| 50 | HNRPU | 16149 | -2.925 | -0.6305 | Yes | ||
| 51 | DHX38 | 16184 | -2.954 | -0.6233 | Yes | ||
| 52 | SF3A3 | 16187 | -2.956 | -0.6144 | Yes | ||
| 53 | RNMT | 16310 | -3.078 | -0.6116 | Yes | ||
| 54 | LSM2 | 16324 | -3.093 | -0.6029 | Yes | ||
| 55 | SFRS5 | 16445 | -3.226 | -0.5996 | Yes | ||
| 56 | SF3A2 | 16513 | -3.324 | -0.5931 | Yes | ||
| 57 | PAPOLA | 16576 | -3.408 | -0.5860 | Yes | ||
| 58 | METTL3 | 16586 | -3.431 | -0.5761 | Yes | ||
| 59 | PPM1G | 16591 | -3.438 | -0.5658 | Yes | ||
| 60 | NXF1 | 16690 | -3.586 | -0.5602 | Yes | ||
| 61 | DHX9 | 16734 | -3.656 | -0.5514 | Yes | ||
| 62 | POLR2A | 16745 | -3.671 | -0.5408 | Yes | ||
| 63 | HNRPA2B1 | 16752 | -3.672 | -0.5299 | Yes | ||
| 64 | SNRPA | 16774 | -3.726 | -0.5197 | Yes | ||
| 65 | PRPF8 | 16791 | -3.754 | -0.5091 | Yes | ||
| 66 | HNRPA3 | 16822 | -3.801 | -0.4992 | Yes | ||
| 67 | SFRS10 | 16834 | -3.818 | -0.4882 | Yes | ||
| 68 | DDX1 | 16939 | -3.983 | -0.4816 | Yes | ||
| 69 | SNRPD3 | 17084 | -4.270 | -0.4764 | Yes | ||
| 70 | HNRPH1 | 17137 | -4.394 | -0.4658 | Yes | ||
| 71 | DHX16 | 17148 | -4.418 | -0.4529 | Yes | ||
| 72 | RBM17 | 17181 | -4.502 | -0.4410 | Yes | ||
| 73 | SNRPD2 | 17243 | -4.639 | -0.4301 | Yes | ||
| 74 | SFRS6 | 17254 | -4.671 | -0.4164 | Yes | ||
| 75 | CPSF1 | 17324 | -4.825 | -0.4055 | Yes | ||
| 76 | RNPS1 | 17326 | -4.839 | -0.3908 | Yes | ||
| 77 | SNRPF | 17457 | -5.204 | -0.3820 | Yes | ||
| 78 | DDX20 | 17463 | -5.217 | -0.3664 | Yes | ||
| 79 | NCBP2 | 17500 | -5.321 | -0.3521 | Yes | ||
| 80 | LSM7 | 17558 | -5.513 | -0.3384 | Yes | ||
| 81 | SF3A1 | 17578 | -5.571 | -0.3225 | Yes | ||
| 82 | SF3B5 | 17597 | -5.622 | -0.3063 | Yes | ||
| 83 | SFRS2 | 17611 | -5.660 | -0.2898 | Yes | ||
| 84 | SNRP70 | 17617 | -5.680 | -0.2728 | Yes | ||
| 85 | SNRPD1 | 17627 | -5.739 | -0.2558 | Yes | ||
| 86 | CSTF3 | 17644 | -5.783 | -0.2390 | Yes | ||
| 87 | CUGBP1 | 17743 | -6.188 | -0.2255 | Yes | ||
| 88 | TXNL4A | 17752 | -6.215 | -0.2070 | Yes | ||
| 89 | SNRPA1 | 17753 | -6.217 | -0.1881 | Yes | ||
| 90 | CPSF4 | 17952 | -7.150 | -0.1770 | Yes | ||
| 91 | PRPF3 | 17953 | -7.160 | -0.1552 | Yes | ||
| 92 | PHF5A | 17973 | -7.243 | -0.1342 | Yes | ||
| 93 | FUSIP1 | 18008 | -7.442 | -0.1134 | Yes | ||
| 94 | U2AF1 | 18107 | -7.981 | -0.0943 | Yes | ||
| 95 | PRPF4 | 18168 | -8.343 | -0.0722 | Yes | ||
| 96 | SNRPB | 18260 | -9.079 | -0.0495 | Yes | ||
| 97 | SRPK1 | 18424 | -10.862 | -0.0252 | Yes | ||
| 98 | XRN2 | 18470 | -11.662 | 0.0079 | Yes |