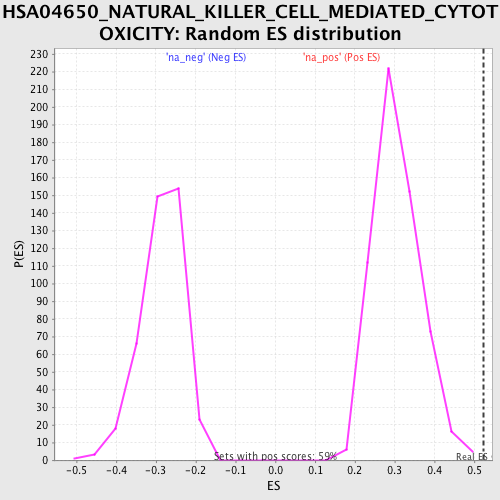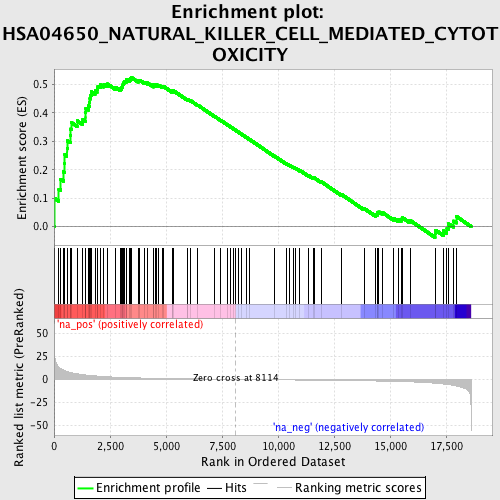
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04650_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY |
| Enrichment Score (ES) | 0.5243393 |
| Normalized Enrichment Score (NES) | 1.7131588 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14772916 |
| FWER p-Value | 0.777 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 12 | 30.494 | 0.0996 | Yes | ||
| 2 | NFATC1 | 213 | 13.199 | 0.1322 | Yes | ||
| 3 | IFNAR2 | 295 | 11.771 | 0.1665 | Yes | ||
| 4 | LAT | 424 | 10.053 | 0.1927 | Yes | ||
| 5 | TNFSF10 | 470 | 9.581 | 0.2217 | Yes | ||
| 6 | VAV1 | 479 | 9.504 | 0.2525 | Yes | ||
| 7 | FYN | 575 | 8.548 | 0.2755 | Yes | ||
| 8 | LCK | 599 | 8.357 | 0.3017 | Yes | ||
| 9 | ZAP70 | 729 | 7.525 | 0.3195 | Yes | ||
| 10 | SH3BP2 | 742 | 7.475 | 0.3434 | Yes | ||
| 11 | LCP2 | 768 | 7.342 | 0.3662 | Yes | ||
| 12 | PIK3CG | 1028 | 6.132 | 0.3724 | Yes | ||
| 13 | PPP3CC | 1281 | 5.185 | 0.3758 | Yes | ||
| 14 | RAC2 | 1399 | 4.842 | 0.3854 | Yes | ||
| 15 | ICAM1 | 1401 | 4.841 | 0.4013 | Yes | ||
| 16 | ITGAL | 1416 | 4.798 | 0.4163 | Yes | ||
| 17 | PPP3CB | 1536 | 4.496 | 0.4247 | Yes | ||
| 18 | KRAS | 1579 | 4.377 | 0.4368 | Yes | ||
| 19 | PIK3CD | 1599 | 4.319 | 0.4500 | Yes | ||
| 20 | NFAT5 | 1621 | 4.255 | 0.4628 | Yes | ||
| 21 | KLRD1 | 1661 | 4.164 | 0.4744 | Yes | ||
| 22 | MAPK3 | 1828 | 3.801 | 0.4779 | Yes | ||
| 23 | IFNGR2 | 1917 | 3.602 | 0.4850 | Yes | ||
| 24 | PTK2B | 1955 | 3.523 | 0.4946 | Yes | ||
| 25 | PAK1 | 2078 | 3.295 | 0.4989 | Yes | ||
| 26 | PRKCB1 | 2223 | 3.074 | 0.5012 | Yes | ||
| 27 | CASP3 | 2379 | 2.804 | 0.5020 | Yes | ||
| 28 | IFNGR1 | 2748 | 2.388 | 0.4900 | Yes | ||
| 29 | CSF2 | 2971 | 2.160 | 0.4851 | Yes | ||
| 30 | SOS2 | 3021 | 2.119 | 0.4895 | Yes | ||
| 31 | PPP3CA | 3049 | 2.097 | 0.4949 | Yes | ||
| 32 | PLCG2 | 3072 | 2.076 | 0.5005 | Yes | ||
| 33 | HCST | 3105 | 2.050 | 0.5055 | Yes | ||
| 34 | PRF1 | 3125 | 2.034 | 0.5112 | Yes | ||
| 35 | FAS | 3215 | 1.961 | 0.5128 | Yes | ||
| 36 | TNFRSF10B | 3231 | 1.951 | 0.5185 | Yes | ||
| 37 | PPP3R1 | 3379 | 1.830 | 0.5165 | Yes | ||
| 38 | ITGB2 | 3410 | 1.810 | 0.5209 | Yes | ||
| 39 | FCER1G | 3455 | 1.782 | 0.5243 | Yes | ||
| 40 | RAC1 | 3748 | 1.593 | 0.5138 | No | ||
| 41 | FCGR3A | 3808 | 1.551 | 0.5157 | No | ||
| 42 | KLRK1 | 4044 | 1.418 | 0.5077 | No | ||
| 43 | KIR3DL1 | 4163 | 1.352 | 0.5058 | No | ||
| 44 | TNF | 4435 | 1.216 | 0.4951 | No | ||
| 45 | PIK3CA | 4439 | 1.215 | 0.4990 | No | ||
| 46 | PIK3R1 | 4537 | 1.166 | 0.4976 | No | ||
| 47 | IFNAR1 | 4558 | 1.157 | 0.5003 | No | ||
| 48 | GRB2 | 4679 | 1.109 | 0.4975 | No | ||
| 49 | IFNA5 | 4851 | 1.042 | 0.4916 | No | ||
| 50 | IFNA13 | 4886 | 1.025 | 0.4932 | No | ||
| 51 | CD247 | 5274 | 0.876 | 0.4752 | No | ||
| 52 | VAV2 | 5303 | 0.865 | 0.4765 | No | ||
| 53 | PIK3R5 | 5314 | 0.861 | 0.4788 | No | ||
| 54 | SHC1 | 5949 | 0.636 | 0.4467 | No | ||
| 55 | SH2D1A | 5955 | 0.634 | 0.4485 | No | ||
| 56 | SOS1 | 6098 | 0.595 | 0.4428 | No | ||
| 57 | IFNB1 | 6408 | 0.497 | 0.4277 | No | ||
| 58 | NFATC3 | 7180 | 0.275 | 0.3870 | No | ||
| 59 | PRKCA | 7416 | 0.203 | 0.3750 | No | ||
| 60 | FASLG | 7752 | 0.101 | 0.3572 | No | ||
| 61 | SHC3 | 7883 | 0.066 | 0.3504 | No | ||
| 62 | GZMB | 8027 | 0.024 | 0.3428 | No | ||
| 63 | RAC3 | 8082 | 0.009 | 0.3399 | No | ||
| 64 | KLRC3 | 8208 | -0.026 | 0.3332 | No | ||
| 65 | TYROBP | 8369 | -0.074 | 0.3248 | No | ||
| 66 | IFNA2 | 8575 | -0.126 | 0.3142 | No | ||
| 67 | NCR1 | 8725 | -0.162 | 0.3067 | No | ||
| 68 | NFATC2 | 9829 | -0.432 | 0.2485 | No | ||
| 69 | CD48 | 10374 | -0.575 | 0.2210 | No | ||
| 70 | ARAF | 10504 | -0.604 | 0.2161 | No | ||
| 71 | IFNA6 | 10685 | -0.646 | 0.2085 | No | ||
| 72 | RAF1 | 10793 | -0.671 | 0.2049 | No | ||
| 73 | NFATC4 | 10972 | -0.710 | 0.1976 | No | ||
| 74 | KLRC2 | 11367 | -0.804 | 0.1790 | No | ||
| 75 | PLCG1 | 11581 | -0.861 | 0.1703 | No | ||
| 76 | IFNA1 | 11606 | -0.870 | 0.1719 | No | ||
| 77 | IFNA7 | 11917 | -0.948 | 0.1583 | No | ||
| 78 | HRAS | 12835 | -1.210 | 0.1127 | No | ||
| 79 | KLRC1 | 13840 | -1.543 | 0.0636 | No | ||
| 80 | MAP2K1 | 14347 | -1.742 | 0.0420 | No | ||
| 81 | PPP3R2 | 14422 | -1.773 | 0.0438 | No | ||
| 82 | SH2D1B | 14444 | -1.784 | 0.0486 | No | ||
| 83 | IFNA14 | 14502 | -1.812 | 0.0514 | No | ||
| 84 | IFNG | 14643 | -1.874 | 0.0500 | No | ||
| 85 | PTPN11 | 15159 | -2.141 | 0.0293 | No | ||
| 86 | IFNA4 | 15379 | -2.268 | 0.0249 | No | ||
| 87 | PIK3R2 | 15490 | -2.342 | 0.0267 | No | ||
| 88 | BID | 15555 | -2.387 | 0.0311 | No | ||
| 89 | MAPK1 | 15894 | -2.679 | 0.0216 | No | ||
| 90 | MAP2K2 | 17005 | -4.113 | -0.0248 | No | ||
| 91 | NRAS | 17023 | -4.143 | -0.0121 | No | ||
| 92 | SYK | 17384 | -4.994 | -0.0151 | No | ||
| 93 | PIK3R3 | 17535 | -5.453 | -0.0053 | No | ||
| 94 | ICAM2 | 17608 | -5.657 | 0.0094 | No | ||
| 95 | PIK3CB | 17817 | -6.520 | 0.0196 | No | ||
| 96 | PTPN6 | 17950 | -7.136 | 0.0360 | No |