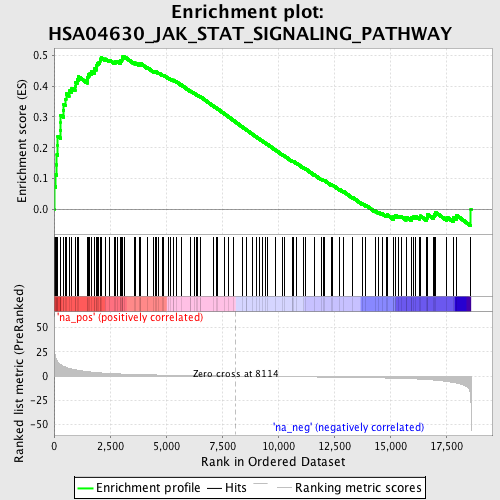
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
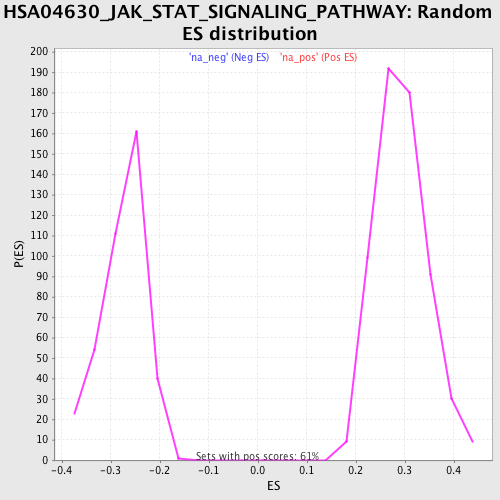
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04630_JAK_STAT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.49489626 |
| Normalized Enrichment Score (NES) | 1.6974632 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15196298 |
| FWER p-Value | 0.835 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STAT4 | 6 | 35.873 | 0.0760 | Yes | ||
| 2 | IL15 | 74 | 18.963 | 0.1127 | Yes | ||
| 3 | IL3RA | 118 | 16.090 | 0.1446 | Yes | ||
| 4 | JAK3 | 126 | 15.779 | 0.1777 | Yes | ||
| 5 | CISH | 161 | 14.580 | 0.2069 | Yes | ||
| 6 | IL28RA | 167 | 14.363 | 0.2372 | Yes | ||
| 7 | PRLR | 283 | 11.918 | 0.2563 | Yes | ||
| 8 | IFNAR2 | 295 | 11.771 | 0.2807 | Yes | ||
| 9 | STAT1 | 300 | 11.691 | 0.3054 | Yes | ||
| 10 | IL5 | 412 | 10.191 | 0.3210 | Yes | ||
| 11 | PIAS3 | 433 | 9.998 | 0.3412 | Yes | ||
| 12 | IL7R | 498 | 9.353 | 0.3577 | Yes | ||
| 13 | SOCS2 | 532 | 8.965 | 0.3749 | Yes | ||
| 14 | JAK2 | 667 | 7.896 | 0.3845 | Yes | ||
| 15 | STAT3 | 790 | 7.215 | 0.3932 | Yes | ||
| 16 | IL12A | 937 | 6.582 | 0.3993 | Yes | ||
| 17 | STAT5A | 947 | 6.564 | 0.4128 | Yes | ||
| 18 | PIK3CG | 1028 | 6.132 | 0.4215 | Yes | ||
| 19 | STAM | 1088 | 5.872 | 0.4308 | Yes | ||
| 20 | IL7 | 1475 | 4.642 | 0.4198 | Yes | ||
| 21 | AKT3 | 1488 | 4.622 | 0.4290 | Yes | ||
| 22 | PIAS1 | 1518 | 4.536 | 0.4370 | Yes | ||
| 23 | PIK3CD | 1599 | 4.319 | 0.4419 | Yes | ||
| 24 | CCND3 | 1660 | 4.168 | 0.4475 | Yes | ||
| 25 | IL10RB | 1791 | 3.890 | 0.4488 | Yes | ||
| 26 | CTF1 | 1810 | 3.845 | 0.4560 | Yes | ||
| 27 | IL21R | 1881 | 3.680 | 0.4600 | Yes | ||
| 28 | SPRED2 | 1896 | 3.640 | 0.4670 | Yes | ||
| 29 | IFNGR2 | 1917 | 3.602 | 0.4736 | Yes | ||
| 30 | BCL2L1 | 1998 | 3.459 | 0.4766 | Yes | ||
| 31 | SOCS3 | 2048 | 3.357 | 0.4811 | Yes | ||
| 32 | STAM2 | 2052 | 3.352 | 0.4880 | Yes | ||
| 33 | CRLF2 | 2102 | 3.264 | 0.4923 | Yes | ||
| 34 | STAT2 | 2298 | 2.933 | 0.4880 | Yes | ||
| 35 | PIM1 | 2474 | 2.686 | 0.4843 | Yes | ||
| 36 | CLCF1 | 2689 | 2.444 | 0.4779 | Yes | ||
| 37 | IFNGR1 | 2748 | 2.388 | 0.4798 | Yes | ||
| 38 | SOCS5 | 2819 | 2.312 | 0.4810 | Yes | ||
| 39 | JAK1 | 2969 | 2.161 | 0.4775 | Yes | ||
| 40 | CSF2 | 2971 | 2.160 | 0.4820 | Yes | ||
| 41 | SOS2 | 3021 | 2.119 | 0.4839 | Yes | ||
| 42 | EPOR | 3032 | 2.109 | 0.4878 | Yes | ||
| 43 | IL13RA1 | 3037 | 2.104 | 0.4921 | Yes | ||
| 44 | CREBBP | 3069 | 2.079 | 0.4948 | Yes | ||
| 45 | CSF2RA | 3148 | 2.015 | 0.4949 | Yes | ||
| 46 | IL6ST | 3579 | 1.698 | 0.4752 | No | ||
| 47 | IL20 | 3649 | 1.659 | 0.4750 | No | ||
| 48 | IL11 | 3819 | 1.542 | 0.4692 | No | ||
| 49 | TSLP | 3836 | 1.535 | 0.4716 | No | ||
| 50 | LIF | 3841 | 1.532 | 0.4746 | No | ||
| 51 | IL2 | 4161 | 1.353 | 0.4602 | No | ||
| 52 | PIK3CA | 4439 | 1.215 | 0.4478 | No | ||
| 53 | PIK3R1 | 4537 | 1.166 | 0.4451 | No | ||
| 54 | IFNAR1 | 4558 | 1.157 | 0.4464 | No | ||
| 55 | GRB2 | 4679 | 1.109 | 0.4423 | No | ||
| 56 | IFNA5 | 4851 | 1.042 | 0.4353 | No | ||
| 57 | IFNA13 | 4886 | 1.025 | 0.4356 | No | ||
| 58 | IL22 | 5108 | 0.940 | 0.4257 | No | ||
| 59 | SPRY2 | 5192 | 0.910 | 0.4231 | No | ||
| 60 | PIK3R5 | 5314 | 0.861 | 0.4184 | No | ||
| 61 | IL13RA2 | 5321 | 0.859 | 0.4199 | No | ||
| 62 | LEPR | 5449 | 0.813 | 0.4147 | No | ||
| 63 | STAT5B | 5666 | 0.724 | 0.4046 | No | ||
| 64 | TPO | 6067 | 0.604 | 0.3842 | No | ||
| 65 | SOS1 | 6098 | 0.595 | 0.3839 | No | ||
| 66 | IL5RA | 6264 | 0.542 | 0.3761 | No | ||
| 67 | MPL | 6362 | 0.512 | 0.3719 | No | ||
| 68 | IFNB1 | 6408 | 0.497 | 0.3706 | No | ||
| 69 | TYK2 | 6516 | 0.464 | 0.3658 | No | ||
| 70 | GHR | 6541 | 0.456 | 0.3654 | No | ||
| 71 | SPRY1 | 7102 | 0.296 | 0.3358 | No | ||
| 72 | PIAS4 | 7229 | 0.260 | 0.3295 | No | ||
| 73 | IL23A | 7289 | 0.242 | 0.3268 | No | ||
| 74 | CCND2 | 7610 | 0.143 | 0.3098 | No | ||
| 75 | OSMR | 7793 | 0.090 | 0.3002 | No | ||
| 76 | IL23R | 8005 | 0.028 | 0.2888 | No | ||
| 77 | IL4R | 8410 | -0.083 | 0.2671 | No | ||
| 78 | IL12B | 8571 | -0.125 | 0.2587 | No | ||
| 79 | IFNA2 | 8575 | -0.126 | 0.2588 | No | ||
| 80 | SPRY4 | 8864 | -0.194 | 0.2437 | No | ||
| 81 | IL2RG | 9022 | -0.235 | 0.2357 | No | ||
| 82 | EPO | 9168 | -0.269 | 0.2284 | No | ||
| 83 | IL19 | 9304 | -0.305 | 0.2217 | No | ||
| 84 | IL22RA1 | 9439 | -0.335 | 0.2152 | No | ||
| 85 | IL9 | 9510 | -0.349 | 0.2122 | No | ||
| 86 | IFNK | 9894 | -0.451 | 0.1924 | No | ||
| 87 | IL22RA2 | 10192 | -0.526 | 0.1775 | No | ||
| 88 | PIAS2 | 10287 | -0.554 | 0.1735 | No | ||
| 89 | LEP | 10634 | -0.635 | 0.1562 | No | ||
| 90 | IL21 | 10658 | -0.640 | 0.1563 | No | ||
| 91 | IFNA6 | 10685 | -0.646 | 0.1563 | No | ||
| 92 | PRL | 10836 | -0.682 | 0.1496 | No | ||
| 93 | IFNE1 | 11117 | -0.743 | 0.1360 | No | ||
| 94 | LIFR | 11217 | -0.768 | 0.1323 | No | ||
| 95 | IFNA1 | 11606 | -0.870 | 0.1132 | No | ||
| 96 | IFNA7 | 11917 | -0.948 | 0.0984 | No | ||
| 97 | IL12RB2 | 12015 | -0.973 | 0.0952 | No | ||
| 98 | SOCS7 | 12051 | -0.985 | 0.0954 | No | ||
| 99 | CBLC | 12364 | -1.068 | 0.0808 | No | ||
| 100 | CNTFR | 12433 | -1.091 | 0.0795 | No | ||
| 101 | IL4 | 12761 | -1.188 | 0.0643 | No | ||
| 102 | SPRED1 | 12934 | -1.240 | 0.0576 | No | ||
| 103 | IL9R | 13322 | -1.362 | 0.0396 | No | ||
| 104 | AKT1 | 13777 | -1.515 | 0.0182 | No | ||
| 105 | CSF3 | 13921 | -1.574 | 0.0139 | No | ||
| 106 | CSF3R | 14327 | -1.731 | -0.0044 | No | ||
| 107 | IFNA14 | 14502 | -1.812 | -0.0099 | No | ||
| 108 | IFNG | 14643 | -1.874 | -0.0135 | No | ||
| 109 | AKT2 | 14825 | -1.957 | -0.0192 | No | ||
| 110 | IL24 | 14861 | -1.977 | -0.0169 | No | ||
| 111 | STAT6 | 15151 | -2.134 | -0.0279 | No | ||
| 112 | PTPN11 | 15159 | -2.141 | -0.0238 | No | ||
| 113 | IL6 | 15227 | -2.173 | -0.0228 | No | ||
| 114 | CBL | 15237 | -2.177 | -0.0186 | No | ||
| 115 | IFNA4 | 15379 | -2.268 | -0.0214 | No | ||
| 116 | PIK3R2 | 15490 | -2.342 | -0.0224 | No | ||
| 117 | CNTF | 15710 | -2.495 | -0.0290 | No | ||
| 118 | IL13 | 15728 | -2.516 | -0.0245 | No | ||
| 119 | IL6R | 15944 | -2.727 | -0.0304 | No | ||
| 120 | SOCS4 | 15952 | -2.737 | -0.0249 | No | ||
| 121 | IL10 | 16046 | -2.825 | -0.0239 | No | ||
| 122 | IL12RB1 | 16133 | -2.907 | -0.0224 | No | ||
| 123 | IL2RA | 16295 | -3.063 | -0.0246 | No | ||
| 124 | MYC | 16349 | -3.119 | -0.0208 | No | ||
| 125 | IL15RA | 16638 | -3.510 | -0.0290 | No | ||
| 126 | IL2RB | 16652 | -3.529 | -0.0222 | No | ||
| 127 | IL3 | 16663 | -3.549 | -0.0152 | No | ||
| 128 | CBLB | 16935 | -3.978 | -0.0214 | No | ||
| 129 | SOCS1 | 16975 | -4.049 | -0.0149 | No | ||
| 130 | IL10RA | 17036 | -4.183 | -0.0092 | No | ||
| 131 | PIK3R3 | 17535 | -5.453 | -0.0246 | No | ||
| 132 | PIK3CB | 17817 | -6.520 | -0.0259 | No | ||
| 133 | PTPN6 | 17950 | -7.136 | -0.0179 | No | ||
| 134 | CCND1 | 18607 | -25.315 | 0.0005 | No |