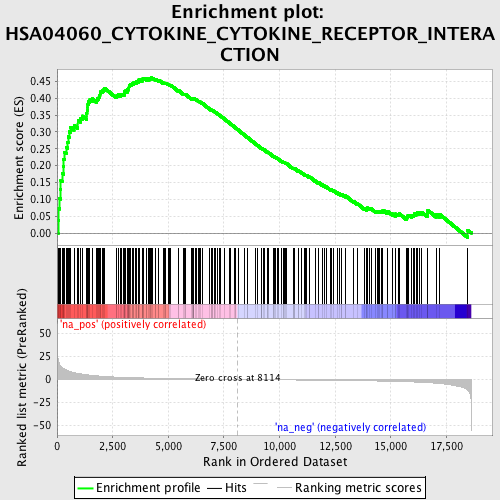
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04060_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION |
| Enrichment Score (ES) | 0.4613379 |
| Normalized Enrichment Score (NES) | 1.6862749 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1514263 |
| FWER p-Value | 0.866 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KIT | 44 | 21.341 | 0.0386 | Yes | ||
| 2 | IL15 | 74 | 18.963 | 0.0735 | Yes | ||
| 3 | IL3RA | 118 | 16.090 | 0.1021 | Yes | ||
| 4 | CD27 | 146 | 15.080 | 0.1296 | Yes | ||
| 5 | IL28RA | 167 | 14.363 | 0.1562 | Yes | ||
| 6 | IL17RA | 228 | 12.828 | 0.1776 | Yes | ||
| 7 | PRLR | 283 | 11.918 | 0.1976 | Yes | ||
| 8 | IFNAR2 | 295 | 11.771 | 0.2196 | Yes | ||
| 9 | TNFRSF18 | 327 | 11.227 | 0.2395 | Yes | ||
| 10 | IL5 | 412 | 10.191 | 0.2545 | Yes | ||
| 11 | TNFSF10 | 470 | 9.581 | 0.2698 | Yes | ||
| 12 | IL7R | 498 | 9.353 | 0.2864 | Yes | ||
| 13 | ACVR1 | 542 | 8.880 | 0.3011 | Yes | ||
| 14 | TNFRSF1A | 593 | 8.427 | 0.3146 | Yes | ||
| 15 | VEGFA | 761 | 7.385 | 0.3197 | Yes | ||
| 16 | IL12A | 937 | 6.582 | 0.3228 | Yes | ||
| 17 | TGFBR1 | 938 | 6.577 | 0.3355 | Yes | ||
| 18 | TNFRSF19 | 1056 | 6.003 | 0.3407 | Yes | ||
| 19 | TGFB1 | 1133 | 5.662 | 0.3474 | Yes | ||
| 20 | TNFSF11 | 1327 | 5.027 | 0.3466 | Yes | ||
| 21 | LTB | 1330 | 5.021 | 0.3561 | Yes | ||
| 22 | CCL27 | 1344 | 4.975 | 0.3650 | Yes | ||
| 23 | CCL4 | 1376 | 4.901 | 0.3727 | Yes | ||
| 24 | TNFSF9 | 1382 | 4.890 | 0.3819 | Yes | ||
| 25 | ACVR1B | 1414 | 4.804 | 0.3894 | Yes | ||
| 26 | IL7 | 1475 | 4.642 | 0.3951 | Yes | ||
| 27 | TNFRSF21 | 1570 | 4.411 | 0.3985 | Yes | ||
| 28 | IL10RB | 1791 | 3.890 | 0.3940 | Yes | ||
| 29 | CTF1 | 1810 | 3.845 | 0.4004 | Yes | ||
| 30 | IL21R | 1881 | 3.680 | 0.4037 | Yes | ||
| 31 | IFNGR2 | 1917 | 3.602 | 0.4087 | Yes | ||
| 32 | CXCR4 | 1928 | 3.584 | 0.4150 | Yes | ||
| 33 | TNFRSF1B | 1966 | 3.503 | 0.4198 | Yes | ||
| 34 | CXCR3 | 2042 | 3.373 | 0.4222 | Yes | ||
| 35 | CRLF2 | 2102 | 3.264 | 0.4252 | Yes | ||
| 36 | CCL25 | 2125 | 3.229 | 0.4303 | Yes | ||
| 37 | IL18RAP | 2667 | 2.466 | 0.4056 | Yes | ||
| 38 | CLCF1 | 2689 | 2.444 | 0.4091 | Yes | ||
| 39 | IFNGR1 | 2748 | 2.388 | 0.4106 | Yes | ||
| 40 | EGF | 2866 | 2.266 | 0.4086 | Yes | ||
| 41 | LTBR | 2894 | 2.237 | 0.4114 | Yes | ||
| 42 | CSF2 | 2971 | 2.160 | 0.4114 | Yes | ||
| 43 | XCL1 | 3013 | 2.126 | 0.4133 | Yes | ||
| 44 | EPOR | 3032 | 2.109 | 0.4164 | Yes | ||
| 45 | IL13RA1 | 3037 | 2.104 | 0.4202 | Yes | ||
| 46 | CXCL12 | 3082 | 2.069 | 0.4218 | Yes | ||
| 47 | CSF2RA | 3148 | 2.015 | 0.4221 | Yes | ||
| 48 | CSF1R | 3150 | 2.015 | 0.4260 | Yes | ||
| 49 | TNFSF13 | 3189 | 1.985 | 0.4277 | Yes | ||
| 50 | FAS | 3215 | 1.961 | 0.4301 | Yes | ||
| 51 | ACVR2A | 3230 | 1.951 | 0.4331 | Yes | ||
| 52 | TNFRSF10B | 3231 | 1.951 | 0.4369 | Yes | ||
| 53 | IL17RB | 3263 | 1.930 | 0.4389 | Yes | ||
| 54 | CCL3 | 3306 | 1.891 | 0.4402 | Yes | ||
| 55 | CX3CR1 | 3366 | 1.840 | 0.4406 | Yes | ||
| 56 | CCL5 | 3378 | 1.830 | 0.4435 | Yes | ||
| 57 | TNFSF14 | 3427 | 1.799 | 0.4443 | Yes | ||
| 58 | CCL28 | 3450 | 1.785 | 0.4466 | Yes | ||
| 59 | FLT1 | 3503 | 1.740 | 0.4471 | Yes | ||
| 60 | IL6ST | 3579 | 1.698 | 0.4463 | Yes | ||
| 61 | TNFRSF14 | 3580 | 1.696 | 0.4495 | Yes | ||
| 62 | TNFRSF11B | 3648 | 1.659 | 0.4491 | Yes | ||
| 63 | IL20 | 3649 | 1.659 | 0.4523 | Yes | ||
| 64 | KITLG | 3672 | 1.641 | 0.4542 | Yes | ||
| 65 | PDGFRB | 3690 | 1.627 | 0.4564 | Yes | ||
| 66 | IL11 | 3819 | 1.542 | 0.4524 | Yes | ||
| 67 | TSLP | 3836 | 1.535 | 0.4545 | Yes | ||
| 68 | LIF | 3841 | 1.532 | 0.4573 | Yes | ||
| 69 | TNFRSF25 | 3875 | 1.511 | 0.4584 | Yes | ||
| 70 | HGF | 3902 | 1.498 | 0.4598 | Yes | ||
| 71 | CXCL5 | 4029 | 1.425 | 0.4557 | Yes | ||
| 72 | PPBP | 4039 | 1.422 | 0.4580 | Yes | ||
| 73 | CCR9 | 4115 | 1.373 | 0.4565 | Yes | ||
| 74 | IL2 | 4161 | 1.353 | 0.4567 | Yes | ||
| 75 | TNFSF13B | 4184 | 1.342 | 0.4581 | Yes | ||
| 76 | IL8RB | 4200 | 1.336 | 0.4598 | Yes | ||
| 77 | CCL7 | 4220 | 1.323 | 0.4613 | Yes | ||
| 78 | CXCL10 | 4293 | 1.281 | 0.4599 | No | ||
| 79 | TNF | 4435 | 1.216 | 0.4546 | No | ||
| 80 | TNFRSF8 | 4437 | 1.215 | 0.4568 | No | ||
| 81 | IFNAR1 | 4558 | 1.157 | 0.4525 | No | ||
| 82 | CXCL2 | 4576 | 1.152 | 0.4538 | No | ||
| 83 | CCL24 | 4772 | 1.073 | 0.4453 | No | ||
| 84 | PDGFB | 4810 | 1.057 | 0.4453 | No | ||
| 85 | IFNA5 | 4851 | 1.042 | 0.4452 | No | ||
| 86 | IFNA13 | 4886 | 1.025 | 0.4453 | No | ||
| 87 | CCR5 | 4992 | 0.981 | 0.4415 | No | ||
| 88 | IL8RA | 5068 | 0.956 | 0.4392 | No | ||
| 89 | IL22 | 5108 | 0.940 | 0.4389 | No | ||
| 90 | LEPR | 5449 | 0.813 | 0.4220 | No | ||
| 91 | INHBB | 5464 | 0.808 | 0.4228 | No | ||
| 92 | IL1B | 5696 | 0.714 | 0.4116 | No | ||
| 93 | INHBC | 5722 | 0.708 | 0.4116 | No | ||
| 94 | BMPR1B | 5756 | 0.695 | 0.4111 | No | ||
| 95 | IL18 | 5768 | 0.692 | 0.4119 | No | ||
| 96 | IL1R2 | 6039 | 0.609 | 0.3984 | No | ||
| 97 | TPO | 6067 | 0.604 | 0.3981 | No | ||
| 98 | MET | 6086 | 0.598 | 0.3982 | No | ||
| 99 | INHBE | 6117 | 0.591 | 0.3977 | No | ||
| 100 | CCL8 | 6121 | 0.587 | 0.3987 | No | ||
| 101 | TNFSF12 | 6124 | 0.585 | 0.3997 | No | ||
| 102 | TNFSF8 | 6132 | 0.584 | 0.4005 | No | ||
| 103 | BMPR1A | 6205 | 0.561 | 0.3976 | No | ||
| 104 | IL5RA | 6264 | 0.542 | 0.3955 | No | ||
| 105 | MPL | 6362 | 0.512 | 0.3912 | No | ||
| 106 | IFNB1 | 6408 | 0.497 | 0.3897 | No | ||
| 107 | CCR8 | 6443 | 0.488 | 0.3888 | No | ||
| 108 | GHR | 6541 | 0.456 | 0.3844 | No | ||
| 109 | CCL19 | 6840 | 0.369 | 0.3690 | No | ||
| 110 | PF4 | 6917 | 0.349 | 0.3655 | No | ||
| 111 | CXCL16 | 6942 | 0.339 | 0.3648 | No | ||
| 112 | EDA | 6988 | 0.326 | 0.3630 | No | ||
| 113 | CD70 | 6998 | 0.324 | 0.3632 | No | ||
| 114 | CCL22 | 7069 | 0.305 | 0.3599 | No | ||
| 115 | EDAR | 7073 | 0.304 | 0.3604 | No | ||
| 116 | BMP7 | 7124 | 0.288 | 0.3582 | No | ||
| 117 | CCR2 | 7204 | 0.268 | 0.3544 | No | ||
| 118 | IL23A | 7289 | 0.242 | 0.3503 | No | ||
| 119 | KDR | 7359 | 0.221 | 0.3470 | No | ||
| 120 | CCL2 | 7511 | 0.172 | 0.3391 | No | ||
| 121 | FASLG | 7752 | 0.101 | 0.3263 | No | ||
| 122 | OSMR | 7793 | 0.090 | 0.3243 | No | ||
| 123 | NGFR | 7964 | 0.043 | 0.3151 | No | ||
| 124 | IL23R | 8005 | 0.028 | 0.3130 | No | ||
| 125 | GDF5 | 8032 | 0.022 | 0.3116 | No | ||
| 126 | IL18R1 | 8154 | -0.010 | 0.3051 | No | ||
| 127 | IL4R | 8410 | -0.083 | 0.2914 | No | ||
| 128 | IL12B | 8571 | -0.125 | 0.2829 | No | ||
| 129 | IFNA2 | 8575 | -0.126 | 0.2830 | No | ||
| 130 | CXCR6 | 8926 | -0.211 | 0.2644 | No | ||
| 131 | IL2RG | 9022 | -0.235 | 0.2597 | No | ||
| 132 | EPO | 9168 | -0.269 | 0.2523 | No | ||
| 133 | ACVR2B | 9196 | -0.276 | 0.2514 | No | ||
| 134 | TGFB2 | 9206 | -0.277 | 0.2514 | No | ||
| 135 | CD40LG | 9288 | -0.301 | 0.2476 | No | ||
| 136 | IL19 | 9304 | -0.305 | 0.2474 | No | ||
| 137 | IL22RA1 | 9439 | -0.335 | 0.2407 | No | ||
| 138 | TNFRSF17 | 9460 | -0.339 | 0.2403 | No | ||
| 139 | IL9 | 9510 | -0.349 | 0.2383 | No | ||
| 140 | FLT3 | 9710 | -0.398 | 0.2282 | No | ||
| 141 | CCR6 | 9732 | -0.406 | 0.2279 | No | ||
| 142 | CCR4 | 9789 | -0.420 | 0.2256 | No | ||
| 143 | EDA2R | 9832 | -0.432 | 0.2242 | No | ||
| 144 | IFNK | 9894 | -0.451 | 0.2217 | No | ||
| 145 | EGFR | 9949 | -0.463 | 0.2197 | No | ||
| 146 | IL1A | 10083 | -0.499 | 0.2134 | No | ||
| 147 | TGFB3 | 10184 | -0.524 | 0.2090 | No | ||
| 148 | IL22RA2 | 10192 | -0.526 | 0.2096 | No | ||
| 149 | CXCL14 | 10218 | -0.532 | 0.2093 | No | ||
| 150 | CXCL9 | 10283 | -0.552 | 0.2069 | No | ||
| 151 | XCR1 | 10305 | -0.558 | 0.2068 | No | ||
| 152 | LTA | 10610 | -0.630 | 0.1915 | No | ||
| 153 | LEP | 10634 | -0.635 | 0.1915 | No | ||
| 154 | IL21 | 10658 | -0.640 | 0.1915 | No | ||
| 155 | IFNA6 | 10685 | -0.646 | 0.1913 | No | ||
| 156 | PRL | 10836 | -0.682 | 0.1845 | No | ||
| 157 | TNFSF18 | 10849 | -0.684 | 0.1851 | No | ||
| 158 | PDGFRA | 10963 | -0.708 | 0.1803 | No | ||
| 159 | IFNE1 | 11117 | -0.743 | 0.1734 | No | ||
| 160 | BMP2 | 11180 | -0.760 | 0.1715 | No | ||
| 161 | LIFR | 11217 | -0.768 | 0.1711 | No | ||
| 162 | AMH | 11336 | -0.796 | 0.1662 | No | ||
| 163 | TNFRSF11A | 11358 | -0.802 | 0.1666 | No | ||
| 164 | IFNA1 | 11606 | -0.870 | 0.1548 | No | ||
| 165 | IL25 | 11746 | -0.902 | 0.1490 | No | ||
| 166 | FLT4 | 11752 | -0.904 | 0.1505 | No | ||
| 167 | IFNA7 | 11917 | -0.948 | 0.1434 | No | ||
| 168 | IL12RB2 | 12015 | -0.973 | 0.1400 | No | ||
| 169 | TNFRSF13B | 12104 | -1.000 | 0.1371 | No | ||
| 170 | CX3CL1 | 12301 | -1.051 | 0.1285 | No | ||
| 171 | TNFRSF9 | 12333 | -1.059 | 0.1288 | No | ||
| 172 | CNTFR | 12433 | -1.091 | 0.1256 | No | ||
| 173 | CXCL13 | 12582 | -1.134 | 0.1197 | No | ||
| 174 | TNFRSF12A | 12672 | -1.163 | 0.1171 | No | ||
| 175 | IL4 | 12761 | -1.188 | 0.1146 | No | ||
| 176 | CCR3 | 12794 | -1.195 | 0.1152 | No | ||
| 177 | CCL1 | 12951 | -1.244 | 0.1091 | No | ||
| 178 | BMPR2 | 12955 | -1.246 | 0.1113 | No | ||
| 179 | IL9R | 13322 | -1.362 | 0.0940 | No | ||
| 180 | PDGFC | 13493 | -1.422 | 0.0875 | No | ||
| 181 | TNFRSF4 | 13837 | -1.542 | 0.0718 | No | ||
| 182 | CXCL11 | 13916 | -1.573 | 0.0706 | No | ||
| 183 | CSF3 | 13921 | -1.574 | 0.0734 | No | ||
| 184 | IL17B | 13952 | -1.587 | 0.0749 | No | ||
| 185 | TNFSF15 | 14047 | -1.618 | 0.0729 | No | ||
| 186 | VEGFC | 14120 | -1.648 | 0.0721 | No | ||
| 187 | CSF3R | 14327 | -1.731 | 0.0642 | No | ||
| 188 | CCL11 | 14383 | -1.755 | 0.0646 | No | ||
| 189 | CSF1 | 14463 | -1.796 | 0.0638 | No | ||
| 190 | IFNA14 | 14502 | -1.812 | 0.0652 | No | ||
| 191 | CXCL1 | 14592 | -1.851 | 0.0639 | No | ||
| 192 | TNFRSF13C | 14634 | -1.869 | 0.0653 | No | ||
| 193 | IFNG | 14643 | -1.874 | 0.0685 | No | ||
| 194 | AMHR2 | 14830 | -1.959 | 0.0621 | No | ||
| 195 | IL24 | 14861 | -1.977 | 0.0643 | No | ||
| 196 | TNFSF4 | 15071 | -2.082 | 0.0569 | No | ||
| 197 | IL1R1 | 15203 | -2.159 | 0.0540 | No | ||
| 198 | IL6 | 15227 | -2.173 | 0.0569 | No | ||
| 199 | TGFBR2 | 15324 | -2.233 | 0.0560 | No | ||
| 200 | IFNA4 | 15379 | -2.268 | 0.0574 | No | ||
| 201 | CNTF | 15710 | -2.495 | 0.0443 | No | ||
| 202 | IL13 | 15728 | -2.516 | 0.0482 | No | ||
| 203 | CD40 | 15769 | -2.557 | 0.0509 | No | ||
| 204 | CCL17 | 15812 | -2.603 | 0.0536 | No | ||
| 205 | IL6R | 15944 | -2.727 | 0.0518 | No | ||
| 206 | IL17A | 16018 | -2.799 | 0.0532 | No | ||
| 207 | IL10 | 16046 | -2.825 | 0.0571 | No | ||
| 208 | IL12RB1 | 16133 | -2.907 | 0.0581 | No | ||
| 209 | IL1RAP | 16186 | -2.955 | 0.0609 | No | ||
| 210 | IL2RA | 16295 | -3.063 | 0.0609 | No | ||
| 211 | CCR1 | 16383 | -3.144 | 0.0622 | No | ||
| 212 | IL15RA | 16638 | -3.510 | 0.0552 | No | ||
| 213 | IL2RB | 16652 | -3.529 | 0.0613 | No | ||
| 214 | IL3 | 16663 | -3.549 | 0.0675 | No | ||
| 215 | IL10RA | 17036 | -4.183 | 0.0554 | No | ||
| 216 | VEGFB | 17189 | -4.511 | 0.0558 | No | ||
| 217 | CCR7 | 18455 | -11.295 | 0.0087 | No |