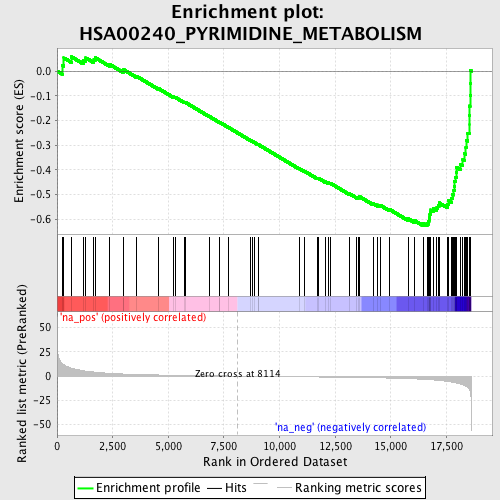
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.62693727 |
| Normalized Enrichment Score (NES) | -2.1196842 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.3273001E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DCK | 229 | 12.823 | 0.0242 | No | ||
| 2 | NP | 306 | 11.619 | 0.0532 | No | ||
| 3 | NT5C2 | 648 | 8.036 | 0.0577 | No | ||
| 4 | NT5M | 1182 | 5.467 | 0.0445 | No | ||
| 5 | ZNRD1 | 1293 | 5.134 | 0.0532 | No | ||
| 6 | RRM2 | 1656 | 4.176 | 0.0456 | No | ||
| 7 | TK1 | 1709 | 4.052 | 0.0543 | No | ||
| 8 | TYMS | 2374 | 2.809 | 0.0265 | No | ||
| 9 | TK2 | 2970 | 2.160 | 0.0006 | No | ||
| 10 | POLD1 | 2998 | 2.135 | 0.0052 | No | ||
| 11 | UPP1 | 3589 | 1.691 | -0.0218 | No | ||
| 12 | NUDT2 | 4569 | 1.154 | -0.0713 | No | ||
| 13 | RRM1 | 5226 | 0.897 | -0.1042 | No | ||
| 14 | ENTPD5 | 5312 | 0.862 | -0.1063 | No | ||
| 15 | POLD4 | 5721 | 0.708 | -0.1263 | No | ||
| 16 | POLA1 | 5762 | 0.693 | -0.1265 | No | ||
| 17 | ENTPD1 | 6852 | 0.367 | -0.1841 | No | ||
| 18 | CTPS2 | 7281 | 0.245 | -0.2065 | No | ||
| 19 | POLR2I | 7301 | 0.237 | -0.2069 | No | ||
| 20 | DPYD | 7710 | 0.114 | -0.2286 | No | ||
| 21 | ENTPD3 | 8684 | -0.151 | -0.2806 | No | ||
| 22 | DPYS | 8802 | -0.181 | -0.2864 | No | ||
| 23 | ITPA | 8871 | -0.196 | -0.2895 | No | ||
| 24 | AICDA | 9029 | -0.237 | -0.2973 | No | ||
| 25 | TXNRD1 | 9056 | -0.242 | -0.2980 | No | ||
| 26 | POLR2C | 10880 | -0.692 | -0.3944 | No | ||
| 27 | POLR2B | 11131 | -0.747 | -0.4057 | No | ||
| 28 | POLD3 | 11705 | -0.892 | -0.4341 | No | ||
| 29 | NT5C3 | 11730 | -0.897 | -0.4328 | No | ||
| 30 | POLE4 | 12046 | -0.983 | -0.4470 | No | ||
| 31 | POLR3K | 12184 | -1.020 | -0.4515 | No | ||
| 32 | POLE2 | 12276 | -1.045 | -0.4534 | No | ||
| 33 | CANT1 | 13143 | -1.304 | -0.4964 | No | ||
| 34 | NT5C1B | 13477 | -1.414 | -0.5103 | No | ||
| 35 | ENTPD8 | 13547 | -1.440 | -0.5100 | No | ||
| 36 | NME4 | 13592 | -1.456 | -0.5082 | No | ||
| 37 | CDA | 14201 | -1.678 | -0.5362 | No | ||
| 38 | POLR2K | 14402 | -1.764 | -0.5420 | No | ||
| 39 | PNPT1 | 14536 | -1.828 | -0.5439 | No | ||
| 40 | RRM2B | 14943 | -2.017 | -0.5601 | No | ||
| 41 | POLR2G | 15779 | -2.565 | -0.5978 | No | ||
| 42 | PRIM1 | 16066 | -2.838 | -0.6051 | No | ||
| 43 | TXNRD2 | 16471 | -3.268 | -0.6176 | Yes | ||
| 44 | ENTPD6 | 16631 | -3.500 | -0.6162 | Yes | ||
| 45 | UPB1 | 16689 | -3.586 | -0.6091 | Yes | ||
| 46 | POLR1D | 16717 | -3.627 | -0.6002 | Yes | ||
| 47 | POLR2A | 16745 | -3.671 | -0.5912 | Yes | ||
| 48 | DTYMK | 16755 | -3.679 | -0.5812 | Yes | ||
| 49 | ECGF1 | 16780 | -3.735 | -0.5719 | Yes | ||
| 50 | NT5E | 16801 | -3.765 | -0.5622 | Yes | ||
| 51 | POLR3A | 16904 | -3.928 | -0.5565 | Yes | ||
| 52 | ENTPD4 | 17071 | -4.242 | -0.5534 | Yes | ||
| 53 | UCK1 | 17120 | -4.370 | -0.5435 | Yes | ||
| 54 | UPP2 | 17171 | -4.473 | -0.5335 | Yes | ||
| 55 | NME6 | 17545 | -5.482 | -0.5380 | Yes | ||
| 56 | NME7 | 17600 | -5.637 | -0.5249 | Yes | ||
| 57 | POLE | 17722 | -6.104 | -0.5140 | Yes | ||
| 58 | POLR2E | 17780 | -6.378 | -0.4989 | Yes | ||
| 59 | POLR2H | 17832 | -6.572 | -0.4829 | Yes | ||
| 60 | POLR2J | 17854 | -6.637 | -0.4651 | Yes | ||
| 61 | UMPS | 17870 | -6.721 | -0.4468 | Yes | ||
| 62 | RFC5 | 17910 | -6.942 | -0.4291 | Yes | ||
| 63 | POLE3 | 17945 | -7.119 | -0.4107 | Yes | ||
| 64 | POLA2 | 17962 | -7.207 | -0.3910 | Yes | ||
| 65 | NT5C | 18127 | -8.085 | -0.3768 | Yes | ||
| 66 | POLD2 | 18238 | -8.926 | -0.3573 | Yes | ||
| 67 | DHODH | 18299 | -9.466 | -0.3336 | Yes | ||
| 68 | NME2 | 18375 | -10.218 | -0.3085 | Yes | ||
| 69 | POLR3H | 18389 | -10.361 | -0.2797 | Yes | ||
| 70 | CAD | 18459 | -11.380 | -0.2510 | Yes | ||
| 71 | NME1 | 18522 | -13.026 | -0.2173 | Yes | ||
| 72 | POLR3B | 18524 | -13.236 | -0.1796 | Yes | ||
| 73 | POLR1A | 18533 | -13.643 | -0.1412 | Yes | ||
| 74 | DCTD | 18561 | -15.395 | -0.0988 | Yes | ||
| 75 | CTPS | 18581 | -17.011 | -0.0513 | Yes | ||
| 76 | POLR1B | 18591 | -18.659 | 0.0013 | Yes |