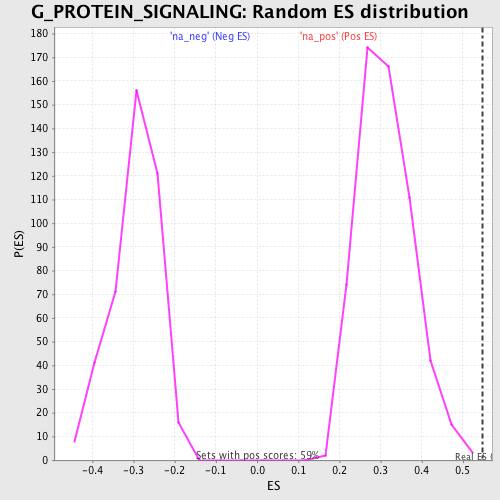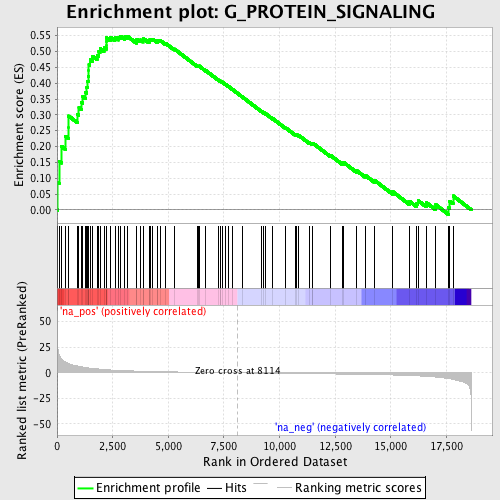
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | G_PROTEIN_SIGNALING |
| Enrichment Score (ES) | 0.54858154 |
| Normalized Enrichment Score (NES) | 1.7546418 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12980597 |
| FWER p-Value | 0.58 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GNA15 | 35 | 22.848 | 0.0872 | Yes | ||
| 2 | PRKCH | 92 | 17.715 | 0.1533 | Yes | ||
| 3 | PRKCQ | 196 | 13.515 | 0.2005 | Yes | ||
| 4 | IL18BP | 371 | 10.713 | 0.2329 | Yes | ||
| 5 | PDE4D | 515 | 9.182 | 0.2610 | Yes | ||
| 6 | PRKACB | 519 | 9.130 | 0.2965 | Yes | ||
| 7 | PRKAR1A | 912 | 6.694 | 0.3014 | Yes | ||
| 8 | GNB2 | 981 | 6.352 | 0.3225 | Yes | ||
| 9 | PDE4A | 1097 | 5.830 | 0.3391 | Yes | ||
| 10 | GNAQ | 1154 | 5.568 | 0.3578 | Yes | ||
| 11 | PPP3CC | 1281 | 5.185 | 0.3712 | Yes | ||
| 12 | PDE4B | 1335 | 5.007 | 0.3879 | Yes | ||
| 13 | CALM3 | 1361 | 4.940 | 0.4058 | Yes | ||
| 14 | ADCY6 | 1400 | 4.842 | 0.4226 | Yes | ||
| 15 | RRAS | 1428 | 4.766 | 0.4398 | Yes | ||
| 16 | GNB5 | 1432 | 4.748 | 0.4581 | Yes | ||
| 17 | PRKACA | 1478 | 4.639 | 0.4738 | Yes | ||
| 18 | KRAS | 1579 | 4.377 | 0.4855 | Yes | ||
| 19 | ADCY9 | 1798 | 3.862 | 0.4888 | Yes | ||
| 20 | PRKCD | 1875 | 3.701 | 0.4991 | Yes | ||
| 21 | SLC9A1 | 1930 | 3.579 | 0.5102 | Yes | ||
| 22 | ITPR1 | 2110 | 3.249 | 0.5132 | Yes | ||
| 23 | PRKD3 | 2213 | 3.090 | 0.5197 | Yes | ||
| 24 | GNG10 | 2217 | 3.082 | 0.5316 | Yes | ||
| 25 | PRKCB1 | 2223 | 3.074 | 0.5433 | Yes | ||
| 26 | KCNJ3 | 2405 | 2.767 | 0.5443 | Yes | ||
| 27 | GNA12 | 2603 | 2.546 | 0.5436 | Yes | ||
| 28 | PLCB3 | 2741 | 2.392 | 0.5456 | Yes | ||
| 29 | GNB1 | 2851 | 2.279 | 0.5486 | Yes | ||
| 30 | PPP3CA | 3049 | 2.097 | 0.5461 | No | ||
| 31 | ARHGEF1 | 3164 | 2.004 | 0.5478 | No | ||
| 32 | GNG12 | 3561 | 1.706 | 0.5331 | No | ||
| 33 | CALM2 | 3582 | 1.696 | 0.5386 | No | ||
| 34 | RHOA | 3738 | 1.602 | 0.5365 | No | ||
| 35 | GNA14 | 3861 | 1.519 | 0.5359 | No | ||
| 36 | AKAP4 | 3893 | 1.502 | 0.5400 | No | ||
| 37 | ADCY5 | 4137 | 1.362 | 0.5322 | No | ||
| 38 | GNAO1 | 4157 | 1.353 | 0.5365 | No | ||
| 39 | GNA13 | 4212 | 1.326 | 0.5388 | No | ||
| 40 | PRKAR1B | 4303 | 1.274 | 0.5389 | No | ||
| 41 | AKAP12 | 4518 | 1.173 | 0.5319 | No | ||
| 42 | AKAP9 | 4527 | 1.169 | 0.5360 | No | ||
| 43 | PDE4C | 4629 | 1.132 | 0.5350 | No | ||
| 44 | AKAP3 | 4862 | 1.037 | 0.5265 | No | ||
| 45 | GNG5 | 5270 | 0.879 | 0.5080 | No | ||
| 46 | PDE1A | 6324 | 0.523 | 0.4532 | No | ||
| 47 | GNG4 | 6360 | 0.512 | 0.4533 | No | ||
| 48 | GNG3 | 6377 | 0.508 | 0.4545 | No | ||
| 49 | PRKD1 | 6687 | 0.418 | 0.4394 | No | ||
| 50 | GNG7 | 7248 | 0.253 | 0.4102 | No | ||
| 51 | PDE8B | 7326 | 0.231 | 0.4069 | No | ||
| 52 | PRKCI | 7357 | 0.222 | 0.4062 | No | ||
| 53 | PRKCA | 7416 | 0.203 | 0.4038 | No | ||
| 54 | PDE1B | 7558 | 0.158 | 0.3968 | No | ||
| 55 | GNGT2 | 7723 | 0.109 | 0.3884 | No | ||
| 56 | GNAZ | 7898 | 0.062 | 0.3793 | No | ||
| 57 | PDE1C | 8334 | -0.064 | 0.3561 | No | ||
| 58 | AKAP10 | 9179 | -0.271 | 0.3116 | No | ||
| 59 | ADCY2 | 9298 | -0.304 | 0.3064 | No | ||
| 60 | PDE7B | 9379 | -0.321 | 0.3033 | No | ||
| 61 | ADCY3 | 9698 | -0.395 | 0.2877 | No | ||
| 62 | GNG13 | 10275 | -0.550 | 0.2588 | No | ||
| 63 | AKAP7 | 10721 | -0.652 | 0.2373 | No | ||
| 64 | AKAP11 | 10739 | -0.657 | 0.2390 | No | ||
| 65 | PDE8A | 10853 | -0.685 | 0.2355 | No | ||
| 66 | GNAI3 | 11323 | -0.793 | 0.2133 | No | ||
| 67 | PRKCE | 11456 | -0.830 | 0.2094 | No | ||
| 68 | ADCY8 | 11499 | -0.842 | 0.2105 | No | ||
| 69 | CALM1 | 12307 | -1.052 | 0.1710 | No | ||
| 70 | HRAS | 12835 | -1.210 | 0.1473 | No | ||
| 71 | GNGT1 | 12864 | -1.221 | 0.1506 | No | ||
| 72 | GNB3 | 13467 | -1.411 | 0.1236 | No | ||
| 73 | PRKCZ | 13858 | -1.555 | 0.1086 | No | ||
| 74 | ADCY4 | 14287 | -1.711 | 0.0922 | No | ||
| 75 | USP5 | 15086 | -2.091 | 0.0573 | No | ||
| 76 | GNA11 | 15833 | -2.621 | 0.0273 | No | ||
| 77 | AKAP8 | 16167 | -2.940 | 0.0208 | No | ||
| 78 | ADCY7 | 16221 | -2.988 | 0.0296 | No | ||
| 79 | PDE7A | 16602 | -3.454 | 0.0225 | No | ||
| 80 | NRAS | 17023 | -4.143 | 0.0160 | No | ||
| 81 | AKAP1 | 17592 | -5.613 | 0.0073 | No | ||
| 82 | PRKAR2B | 17645 | -5.786 | 0.0270 | No | ||
| 83 | PRKAR2A | 17801 | -6.482 | 0.0440 | No |