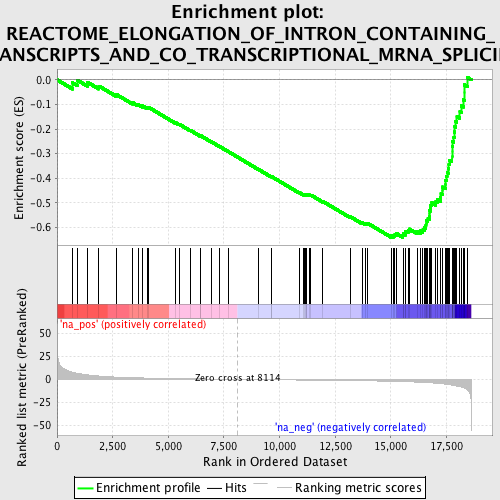
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_ELONGATION_OF_INTRON_CONTAINING_TRANSCRIPTS_AND_CO_TRANSCRIPTIONAL_MRNA_SPLICING |
| Enrichment Score (ES) | -0.6404309 |
| Normalized Enrichment Score (NES) | -2.1809967 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ERCC2 | 697 | 7.736 | -0.0122 | No | ||
| 2 | TCEB2 | 917 | 6.682 | -0.0020 | No | ||
| 3 | RBMX | 1359 | 4.944 | -0.0095 | No | ||
| 4 | CD2BP2 | 1878 | 3.694 | -0.0253 | No | ||
| 5 | PCBP2 | 2658 | 2.477 | -0.0592 | No | ||
| 6 | RBM5 | 3387 | 1.824 | -0.0925 | No | ||
| 7 | ELL | 3645 | 1.660 | -0.1009 | No | ||
| 8 | TCEB3 | 3834 | 1.537 | -0.1060 | No | ||
| 9 | SFRS7 | 4053 | 1.412 | -0.1131 | No | ||
| 10 | CCNT1 | 4108 | 1.376 | -0.1115 | No | ||
| 11 | SUPT4H1 | 5308 | 0.863 | -0.1734 | No | ||
| 12 | SFRS3 | 5496 | 0.795 | -0.1808 | No | ||
| 13 | CSTF2 | 6000 | 0.620 | -0.2059 | No | ||
| 14 | CCNT2 | 6436 | 0.490 | -0.2278 | No | ||
| 15 | SF4 | 6456 | 0.483 | -0.2272 | No | ||
| 16 | SF3B1 | 6934 | 0.342 | -0.2519 | No | ||
| 17 | POLR2I | 7301 | 0.237 | -0.2708 | No | ||
| 18 | PCF11 | 7692 | 0.121 | -0.2915 | No | ||
| 19 | PABPN1 | 9064 | -0.243 | -0.3647 | No | ||
| 20 | SNRPE | 9624 | -0.376 | -0.3936 | No | ||
| 21 | CCAR1 | 9652 | -0.383 | -0.3938 | No | ||
| 22 | POLR2C | 10880 | -0.692 | -0.4577 | No | ||
| 23 | TCEB1 | 11078 | -0.734 | -0.4659 | No | ||
| 24 | POLR2B | 11131 | -0.747 | -0.4663 | No | ||
| 25 | SNRPB2 | 11186 | -0.762 | -0.4667 | No | ||
| 26 | SFRS1 | 11226 | -0.769 | -0.4663 | No | ||
| 27 | MNAT1 | 11230 | -0.770 | -0.4639 | No | ||
| 28 | SRRM1 | 11348 | -0.799 | -0.4676 | No | ||
| 29 | DNAJC8 | 11408 | -0.815 | -0.4681 | No | ||
| 30 | SF3B2 | 11949 | -0.956 | -0.4941 | No | ||
| 31 | FUS | 13173 | -1.312 | -0.5558 | No | ||
| 32 | SF3B4 | 13725 | -1.497 | -0.5806 | No | ||
| 33 | CDK7 | 13859 | -1.555 | -0.5826 | No | ||
| 34 | CSTF1 | 13963 | -1.592 | -0.5830 | No | ||
| 35 | SMC1A | 15029 | -2.063 | -0.6336 | Yes | ||
| 36 | U2AF2 | 15127 | -2.117 | -0.6319 | Yes | ||
| 37 | SFRS9 | 15184 | -2.148 | -0.6279 | Yes | ||
| 38 | NUDT21 | 15254 | -2.189 | -0.6244 | Yes | ||
| 39 | PTBP1 | 15547 | -2.383 | -0.6323 | Yes | ||
| 40 | RDBP | 15564 | -2.391 | -0.6253 | Yes | ||
| 41 | SNRPG | 15665 | -2.464 | -0.6226 | Yes | ||
| 42 | CCNH | 15666 | -2.465 | -0.6145 | Yes | ||
| 43 | POLR2G | 15779 | -2.565 | -0.6121 | Yes | ||
| 44 | MAGOH | 15827 | -2.618 | -0.6060 | Yes | ||
| 45 | SF3A3 | 16187 | -2.956 | -0.6157 | Yes | ||
| 46 | THOC4 | 16343 | -3.113 | -0.6138 | Yes | ||
| 47 | SFRS5 | 16445 | -3.226 | -0.6086 | Yes | ||
| 48 | SF3A2 | 16513 | -3.324 | -0.6013 | Yes | ||
| 49 | PAPOLA | 16576 | -3.408 | -0.5934 | Yes | ||
| 50 | NFX1 | 16582 | -3.424 | -0.5824 | Yes | ||
| 51 | METTL3 | 16586 | -3.431 | -0.5713 | Yes | ||
| 52 | CDK9 | 16670 | -3.556 | -0.5641 | Yes | ||
| 53 | DHX9 | 16734 | -3.656 | -0.5555 | Yes | ||
| 54 | POLR2A | 16745 | -3.671 | -0.5439 | Yes | ||
| 55 | PCBP1 | 16747 | -3.671 | -0.5319 | Yes | ||
| 56 | SNRPA | 16774 | -3.726 | -0.5211 | Yes | ||
| 57 | PRPF8 | 16791 | -3.754 | -0.5096 | Yes | ||
| 58 | ERCC3 | 16844 | -3.832 | -0.4998 | Yes | ||
| 59 | GTF2F1 | 17026 | -4.152 | -0.4959 | Yes | ||
| 60 | SNRPD3 | 17084 | -4.270 | -0.4849 | Yes | ||
| 61 | SNRPD2 | 17243 | -4.639 | -0.4782 | Yes | ||
| 62 | SFRS6 | 17254 | -4.671 | -0.4633 | Yes | ||
| 63 | CPSF1 | 17324 | -4.825 | -0.4512 | Yes | ||
| 64 | RNPS1 | 17326 | -4.839 | -0.4353 | Yes | ||
| 65 | SNRPF | 17457 | -5.204 | -0.4252 | Yes | ||
| 66 | GTF2H1 | 17462 | -5.217 | -0.4083 | Yes | ||
| 67 | NCBP2 | 17500 | -5.321 | -0.3928 | Yes | ||
| 68 | SF3B3 | 17528 | -5.426 | -0.3764 | Yes | ||
| 69 | SF3A1 | 17578 | -5.571 | -0.3607 | Yes | ||
| 70 | SFRS2 | 17611 | -5.660 | -0.3438 | Yes | ||
| 71 | CSTF3 | 17644 | -5.783 | -0.3265 | Yes | ||
| 72 | TH1L | 17749 | -6.207 | -0.3117 | Yes | ||
| 73 | TXNL4A | 17752 | -6.215 | -0.2913 | Yes | ||
| 74 | SNRPA1 | 17753 | -6.217 | -0.2709 | Yes | ||
| 75 | POLR2E | 17780 | -6.378 | -0.2513 | Yes | ||
| 76 | POLR2H | 17832 | -6.572 | -0.2324 | Yes | ||
| 77 | POLR2J | 17854 | -6.637 | -0.2117 | Yes | ||
| 78 | GTF2H3 | 17877 | -6.742 | -0.1908 | Yes | ||
| 79 | YBX1 | 17915 | -6.949 | -0.1699 | Yes | ||
| 80 | PHF5A | 17973 | -7.243 | -0.1491 | Yes | ||
| 81 | U2AF1 | 18107 | -7.981 | -0.1301 | Yes | ||
| 82 | PRPF4 | 18168 | -8.343 | -0.1058 | Yes | ||
| 83 | SNRPB | 18260 | -9.079 | -0.0809 | Yes | ||
| 84 | GTF2H2 | 18309 | -9.581 | -0.0520 | Yes | ||
| 85 | EFTUD2 | 18310 | -9.584 | -0.0204 | Yes | ||
| 86 | GTF2F2 | 18450 | -11.212 | 0.0090 | Yes |
