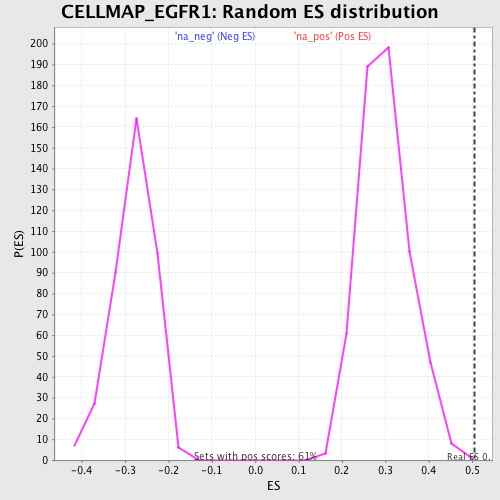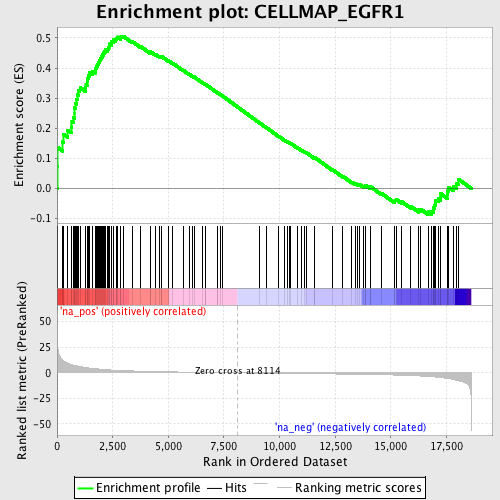
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CELLMAP_EGFR1 |
| Enrichment Score (ES) | 0.5059425 |
| Normalized Enrichment Score (NES) | 1.6925979 |
| Nominal p-value | 0.0016474464 |
| FDR q-value | 0.18556106 |
| FWER p-Value | 0.954 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 12 | 30.494 | 0.0728 | Yes | ||
| 2 | GAB1 | 22 | 26.495 | 0.1361 | Yes | ||
| 3 | GRB7 | 241 | 12.611 | 0.1547 | Yes | ||
| 4 | STAT1 | 300 | 11.691 | 0.1797 | Yes | ||
| 5 | VAV1 | 479 | 9.504 | 0.1929 | Yes | ||
| 6 | SH3KBP1 | 633 | 8.165 | 0.2043 | Yes | ||
| 7 | JAK2 | 667 | 7.896 | 0.2216 | Yes | ||
| 8 | RPS6KA2 | 735 | 7.507 | 0.2360 | Yes | ||
| 9 | STAT3 | 790 | 7.215 | 0.2505 | Yes | ||
| 10 | SMAD3 | 794 | 7.202 | 0.2676 | Yes | ||
| 11 | RALBP1 | 832 | 7.008 | 0.2825 | Yes | ||
| 12 | SH3BGRL | 872 | 6.868 | 0.2970 | Yes | ||
| 13 | PRKAR1A | 912 | 6.694 | 0.3110 | Yes | ||
| 14 | STAT5A | 947 | 6.564 | 0.3249 | Yes | ||
| 15 | PIK3CG | 1028 | 6.132 | 0.3354 | Yes | ||
| 16 | SH3GL3 | 1266 | 5.222 | 0.3351 | Yes | ||
| 17 | RIPK1 | 1297 | 5.115 | 0.3458 | Yes | ||
| 18 | MTA2 | 1349 | 4.965 | 0.3550 | Yes | ||
| 19 | MAPK14 | 1366 | 4.922 | 0.3660 | Yes | ||
| 20 | CAV2 | 1405 | 4.826 | 0.3756 | Yes | ||
| 21 | NCK2 | 1456 | 4.693 | 0.3842 | Yes | ||
| 22 | KRAS | 1579 | 4.377 | 0.3881 | Yes | ||
| 23 | RASA1 | 1707 | 4.056 | 0.3910 | Yes | ||
| 24 | TNIP1 | 1736 | 3.985 | 0.3991 | Yes | ||
| 25 | ABI1 | 1770 | 3.925 | 0.4068 | Yes | ||
| 26 | MAPK3 | 1828 | 3.801 | 0.4128 | Yes | ||
| 27 | CRKL | 1876 | 3.698 | 0.4192 | Yes | ||
| 28 | MAP3K4 | 1905 | 3.628 | 0.4264 | Yes | ||
| 29 | PTK2B | 1955 | 3.523 | 0.4323 | Yes | ||
| 30 | PLSCR1 | 1999 | 3.457 | 0.4383 | Yes | ||
| 31 | SOCS3 | 2048 | 3.357 | 0.4437 | Yes | ||
| 32 | PAK1 | 2078 | 3.295 | 0.4501 | Yes | ||
| 33 | WASL | 2116 | 3.241 | 0.4559 | Yes | ||
| 34 | PLEC1 | 2177 | 3.144 | 0.4602 | Yes | ||
| 35 | ELK4 | 2285 | 2.959 | 0.4616 | Yes | ||
| 36 | STAT2 | 2298 | 2.933 | 0.4680 | Yes | ||
| 37 | GJA1 | 2333 | 2.878 | 0.4731 | Yes | ||
| 38 | PLD2 | 2336 | 2.863 | 0.4799 | Yes | ||
| 39 | JUN | 2446 | 2.711 | 0.4805 | Yes | ||
| 40 | YWHAB | 2453 | 2.702 | 0.4867 | Yes | ||
| 41 | RALGDS | 2518 | 2.641 | 0.4896 | Yes | ||
| 42 | MAP3K14 | 2522 | 2.637 | 0.4958 | Yes | ||
| 43 | HDAC1 | 2646 | 2.493 | 0.4951 | Yes | ||
| 44 | SHOC2 | 2675 | 2.460 | 0.4995 | Yes | ||
| 45 | PLD1 | 2719 | 2.413 | 0.5030 | Yes | ||
| 46 | EGF | 2866 | 2.266 | 0.5006 | Yes | ||
| 47 | HIP1 | 2869 | 2.265 | 0.5059 | Yes | ||
| 48 | JAK1 | 2969 | 2.161 | 0.5058 | No | ||
| 49 | EEF1A1 | 3386 | 1.825 | 0.4877 | No | ||
| 50 | RAC1 | 3748 | 1.593 | 0.4720 | No | ||
| 51 | CREB1 | 4196 | 1.337 | 0.4511 | No | ||
| 52 | ATF1 | 4203 | 1.334 | 0.4540 | No | ||
| 53 | PIK3CA | 4439 | 1.215 | 0.4442 | No | ||
| 54 | GRB14 | 4588 | 1.148 | 0.4390 | No | ||
| 55 | GRB2 | 4679 | 1.109 | 0.4368 | No | ||
| 56 | SMAD2 | 4698 | 1.102 | 0.4384 | No | ||
| 57 | PITPNA | 4984 | 0.984 | 0.4254 | No | ||
| 58 | SPRY2 | 5192 | 0.910 | 0.4164 | No | ||
| 59 | STAT5B | 5666 | 0.724 | 0.3926 | No | ||
| 60 | SHC1 | 5949 | 0.636 | 0.3789 | No | ||
| 61 | SOS1 | 6098 | 0.595 | 0.3723 | No | ||
| 62 | DNM1 | 6160 | 0.575 | 0.3704 | No | ||
| 63 | SP1 | 6515 | 0.465 | 0.3524 | No | ||
| 64 | PTK6 | 6653 | 0.427 | 0.3460 | No | ||
| 65 | PRKD1 | 6687 | 0.418 | 0.3452 | No | ||
| 66 | CSK | 7206 | 0.268 | 0.3179 | No | ||
| 67 | FOS | 7226 | 0.262 | 0.3175 | No | ||
| 68 | PRKCI | 7357 | 0.222 | 0.3110 | No | ||
| 69 | PRKCA | 7416 | 0.203 | 0.3083 | No | ||
| 70 | CAV1 | 9097 | -0.251 | 0.2181 | No | ||
| 71 | AP2A1 | 9402 | -0.327 | 0.2025 | No | ||
| 72 | EGFR | 9949 | -0.463 | 0.1741 | No | ||
| 73 | EPS15 | 10230 | -0.536 | 0.1602 | No | ||
| 74 | RAB5A | 10363 | -0.572 | 0.1545 | No | ||
| 75 | SRC | 10441 | -0.590 | 0.1517 | No | ||
| 76 | ARAF | 10504 | -0.604 | 0.1498 | No | ||
| 77 | RAF1 | 10793 | -0.671 | 0.1359 | No | ||
| 78 | KLF11 | 10976 | -0.711 | 0.1278 | No | ||
| 79 | RPS6KA1 | 11121 | -0.744 | 0.1218 | No | ||
| 80 | NCK1 | 11188 | -0.762 | 0.1200 | No | ||
| 81 | PLCG1 | 11581 | -0.861 | 0.1009 | No | ||
| 82 | SIN3A | 11586 | -0.863 | 0.1028 | No | ||
| 83 | CBLC | 12364 | -1.068 | 0.0634 | No | ||
| 84 | HRAS | 12835 | -1.210 | 0.0409 | No | ||
| 85 | SNCA | 13253 | -1.339 | 0.0215 | No | ||
| 86 | ELF3 | 13432 | -1.399 | 0.0153 | No | ||
| 87 | RGS16 | 13520 | -1.431 | 0.0140 | No | ||
| 88 | ARF4 | 13597 | -1.458 | 0.0134 | No | ||
| 89 | AKT1 | 13777 | -1.515 | 0.0074 | No | ||
| 90 | PRKCZ | 13858 | -1.555 | 0.0068 | No | ||
| 91 | CEBPB | 13876 | -1.561 | 0.0097 | No | ||
| 92 | MCF2 | 14064 | -1.625 | 0.0035 | No | ||
| 93 | MAPK8 | 14071 | -1.628 | 0.0071 | No | ||
| 94 | MAP3K2 | 14560 | -1.841 | -0.0149 | No | ||
| 95 | PTPN11 | 15159 | -2.141 | -0.0420 | No | ||
| 96 | CDC42 | 15186 | -2.149 | -0.0383 | No | ||
| 97 | CBL | 15237 | -2.177 | -0.0357 | No | ||
| 98 | PIK3R2 | 15490 | -2.342 | -0.0437 | No | ||
| 99 | MAPK7 | 15883 | -2.664 | -0.0585 | No | ||
| 100 | PKN2 | 16250 | -3.022 | -0.0710 | No | ||
| 101 | MYC | 16349 | -3.119 | -0.0688 | No | ||
| 102 | RALB | 16672 | -3.562 | -0.0776 | No | ||
| 103 | CRK | 16826 | -3.808 | -0.0767 | No | ||
| 104 | HAT1 | 16908 | -3.934 | -0.0716 | No | ||
| 105 | CBLB | 16935 | -3.978 | -0.0635 | No | ||
| 106 | SOCS1 | 16975 | -4.049 | -0.0558 | No | ||
| 107 | MAP2K2 | 17005 | -4.113 | -0.0475 | No | ||
| 108 | NRAS | 17023 | -4.143 | -0.0384 | No | ||
| 109 | INPPL1 | 17135 | -4.392 | -0.0338 | No | ||
| 110 | CASP9 | 17236 | -4.631 | -0.0281 | No | ||
| 111 | SNRPD2 | 17243 | -4.639 | -0.0173 | No | ||
| 112 | PIK3R3 | 17535 | -5.453 | -0.0199 | No | ||
| 113 | STXBP1 | 17564 | -5.536 | -0.0080 | No | ||
| 114 | RBBP7 | 17593 | -5.614 | 0.0040 | No | ||
| 115 | PIK3CB | 17817 | -6.520 | 0.0076 | No | ||
| 116 | PTPN6 | 17950 | -7.136 | 0.0177 | No | ||
| 117 | TNK2 | 18033 | -7.596 | 0.0315 | No |