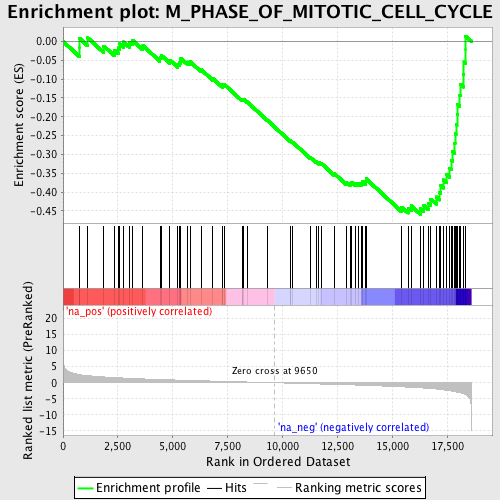
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | M_PHASE_OF_MITOTIC_CELL_CYCLE |
| Enrichment Score (ES) | -0.45931503 |
| Normalized Enrichment Score (NES) | -1.8388493 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06410906 |
| FWER p-Value | 0.706 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PAM | 751 | 2.532 | -0.0160 | No | ||
| 2 | CHMP1A | 754 | 2.528 | 0.0084 | No | ||
| 3 | MAD2L1 | 1128 | 2.202 | 0.0095 | No | ||
| 4 | CCNA2 | 1853 | 1.799 | -0.0121 | No | ||
| 5 | NPM2 | 2336 | 1.598 | -0.0226 | No | ||
| 6 | NUSAP1 | 2509 | 1.543 | -0.0170 | No | ||
| 7 | PIN1 | 2583 | 1.520 | -0.0062 | No | ||
| 8 | TTN | 2754 | 1.461 | -0.0012 | No | ||
| 9 | RAD17 | 3013 | 1.378 | -0.0018 | No | ||
| 10 | SUGT1 | 3148 | 1.333 | 0.0039 | No | ||
| 11 | ATM | 3638 | 1.186 | -0.0110 | No | ||
| 12 | ZWINT | 4415 | 1.000 | -0.0432 | No | ||
| 13 | ANAPC10 | 4467 | 0.987 | -0.0364 | No | ||
| 14 | TGFA | 4867 | 0.900 | -0.0492 | No | ||
| 15 | NDC80 | 5232 | 0.826 | -0.0608 | No | ||
| 16 | CHFR | 5301 | 0.811 | -0.0566 | No | ||
| 17 | CDC23 | 5353 | 0.800 | -0.0516 | No | ||
| 18 | NEK2 | 5362 | 0.796 | -0.0444 | No | ||
| 19 | ANAPC11 | 5680 | 0.730 | -0.0544 | No | ||
| 20 | RCC1 | 5794 | 0.704 | -0.0537 | No | ||
| 21 | ANLN | 6293 | 0.612 | -0.0746 | No | ||
| 22 | CDKN2B | 6825 | 0.514 | -0.0983 | No | ||
| 23 | RAN | 7264 | 0.422 | -0.1178 | No | ||
| 24 | UBE2C | 7270 | 0.421 | -0.1140 | No | ||
| 25 | MAD2L2 | 7364 | 0.408 | -0.1151 | No | ||
| 26 | PRMT5 | 8157 | 0.265 | -0.1552 | No | ||
| 27 | ANAPC4 | 8162 | 0.265 | -0.1529 | No | ||
| 28 | BUB1 | 8225 | 0.254 | -0.1538 | No | ||
| 29 | CDCA5 | 8391 | 0.226 | -0.1605 | No | ||
| 30 | AURKA | 9293 | 0.067 | -0.2084 | No | ||
| 31 | NEK6 | 10344 | -0.127 | -0.2638 | No | ||
| 32 | CENPE | 10373 | -0.134 | -0.2640 | No | ||
| 33 | CDC16 | 10470 | -0.152 | -0.2677 | No | ||
| 34 | CD28 | 11286 | -0.304 | -0.3087 | No | ||
| 35 | CETN1 | 11531 | -0.350 | -0.3185 | No | ||
| 36 | KIF11 | 11659 | -0.378 | -0.3217 | No | ||
| 37 | BUB1B | 11770 | -0.400 | -0.3238 | No | ||
| 38 | BIRC5 | 12364 | -0.512 | -0.3508 | No | ||
| 39 | EPGN | 12897 | -0.619 | -0.3735 | No | ||
| 40 | TTK | 13086 | -0.658 | -0.3773 | No | ||
| 41 | EGF | 13134 | -0.666 | -0.3733 | No | ||
| 42 | TGFB1 | 13308 | -0.704 | -0.3759 | No | ||
| 43 | SSSCA1 | 13452 | -0.737 | -0.3764 | No | ||
| 44 | AKAP8 | 13583 | -0.766 | -0.3760 | No | ||
| 45 | DCTN3 | 13654 | -0.783 | -0.3722 | No | ||
| 46 | KIF15 | 13789 | -0.817 | -0.3716 | No | ||
| 47 | SMC4 | 13804 | -0.821 | -0.3644 | No | ||
| 48 | KNTC1 | 15435 | -1.269 | -0.4400 | No | ||
| 49 | SMC3 | 15722 | -1.364 | -0.4422 | No | ||
| 50 | ZW10 | 15866 | -1.415 | -0.4362 | No | ||
| 51 | TARDBP | 16295 | -1.582 | -0.4440 | Yes | ||
| 52 | CDC25B | 16441 | -1.655 | -0.4358 | Yes | ||
| 53 | NCAPH | 16656 | -1.758 | -0.4303 | Yes | ||
| 54 | PCBP4 | 16759 | -1.815 | -0.4183 | Yes | ||
| 55 | CDC25C | 17009 | -1.959 | -0.4128 | Yes | ||
| 56 | PPP5C | 17161 | -2.096 | -0.4006 | Yes | ||
| 57 | EREG | 17216 | -2.137 | -0.3829 | Yes | ||
| 58 | CIT | 17333 | -2.227 | -0.3676 | Yes | ||
| 59 | TPX2 | 17473 | -2.353 | -0.3523 | Yes | ||
| 60 | ANAPC5 | 17619 | -2.499 | -0.3359 | Yes | ||
| 61 | KIF22 | 17690 | -2.585 | -0.3147 | Yes | ||
| 62 | NOLC1 | 17748 | -2.639 | -0.2922 | Yes | ||
| 63 | PLK1 | 17847 | -2.764 | -0.2708 | Yes | ||
| 64 | NUMA1 | 17864 | -2.791 | -0.2446 | Yes | ||
| 65 | DDX11 | 17909 | -2.844 | -0.2195 | Yes | ||
| 66 | CLIP1 | 17953 | -2.913 | -0.1936 | Yes | ||
| 67 | NBN | 17973 | -2.942 | -0.1662 | Yes | ||
| 68 | PML | 18083 | -3.084 | -0.1422 | Yes | ||
| 69 | MPHOSPH6 | 18116 | -3.151 | -0.1135 | Yes | ||
| 70 | DCTN2 | 18256 | -3.458 | -0.0875 | Yes | ||
| 71 | DLG7 | 18270 | -3.498 | -0.0543 | Yes | ||
| 72 | KIF2C | 18349 | -3.764 | -0.0221 | Yes | ||
| 73 | SMC1A | 18351 | -3.769 | 0.0143 | Yes |