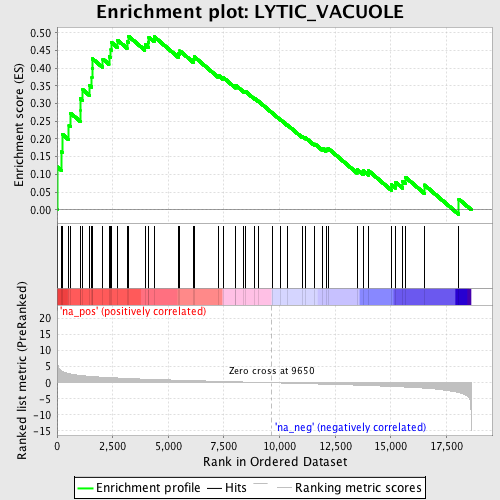
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | LYTIC_VACUOLE |
| Enrichment Score (ES) | 0.49043155 |
| Normalized Enrichment Score (NES) | 1.8749964 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07807851 |
| FWER p-Value | 0.606 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IFI30 | 8 | 8.539 | 0.1212 | Yes | ||
| 2 | CORO1A | 204 | 3.720 | 0.1636 | Yes | ||
| 3 | BCL10 | 229 | 3.599 | 0.2136 | Yes | ||
| 4 | VAMP4 | 532 | 2.844 | 0.2378 | Yes | ||
| 5 | RAB7A | 598 | 2.733 | 0.2732 | Yes | ||
| 6 | TPP1 | 1039 | 2.265 | 0.2818 | Yes | ||
| 7 | VPS45 | 1043 | 2.263 | 0.3138 | Yes | ||
| 8 | USP4 | 1155 | 2.187 | 0.3390 | Yes | ||
| 9 | LAMP2 | 1455 | 1.993 | 0.3512 | Yes | ||
| 10 | CLN5 | 1564 | 1.937 | 0.3730 | Yes | ||
| 11 | CTSC | 1572 | 1.933 | 0.4001 | Yes | ||
| 12 | IL8RB | 1593 | 1.920 | 0.4264 | Yes | ||
| 13 | CD63 | 2057 | 1.701 | 0.4257 | Yes | ||
| 14 | AP3M1 | 2333 | 1.600 | 0.4337 | Yes | ||
| 15 | NEU4 | 2419 | 1.572 | 0.4515 | Yes | ||
| 16 | LAMP3 | 2435 | 1.566 | 0.4730 | Yes | ||
| 17 | GALC | 2722 | 1.478 | 0.4786 | Yes | ||
| 18 | NAGLU | 3146 | 1.334 | 0.4748 | Yes | ||
| 19 | KCNE1 | 3205 | 1.316 | 0.4904 | Yes | ||
| 20 | HPS4 | 3950 | 1.104 | 0.4661 | No | ||
| 21 | MPO | 4100 | 1.072 | 0.4733 | No | ||
| 22 | SPACA3 | 4123 | 1.065 | 0.4873 | No | ||
| 23 | GLA | 4364 | 1.013 | 0.4888 | No | ||
| 24 | TRIM23 | 5456 | 0.777 | 0.4411 | No | ||
| 25 | RAB9A | 5519 | 0.764 | 0.4486 | No | ||
| 26 | USP5 | 6142 | 0.639 | 0.4242 | No | ||
| 27 | TIAL1 | 6154 | 0.637 | 0.4327 | No | ||
| 28 | TYR | 7272 | 0.421 | 0.3785 | No | ||
| 29 | NPC2 | 7468 | 0.389 | 0.3735 | No | ||
| 30 | LRP2 | 8023 | 0.287 | 0.3478 | No | ||
| 31 | DNASE2 | 8031 | 0.284 | 0.3514 | No | ||
| 32 | TOM1L1 | 8390 | 0.227 | 0.3354 | No | ||
| 33 | CTNS | 8474 | 0.212 | 0.3339 | No | ||
| 34 | RAMP2 | 8878 | 0.138 | 0.3142 | No | ||
| 35 | ADRB2 | 9036 | 0.111 | 0.3073 | No | ||
| 36 | RAMP3 | 9670 | -0.004 | 0.2733 | No | ||
| 37 | MYO7A | 10049 | -0.077 | 0.2540 | No | ||
| 38 | CTSF | 10352 | -0.129 | 0.2396 | No | ||
| 39 | SFTPD | 11032 | -0.252 | 0.2066 | No | ||
| 40 | KCNE2 | 11158 | -0.279 | 0.2038 | No | ||
| 41 | ACR | 11589 | -0.361 | 0.1858 | No | ||
| 42 | RAB14 | 11937 | -0.430 | 0.1732 | No | ||
| 43 | CYLC1 | 12087 | -0.457 | 0.1717 | No | ||
| 44 | STS | 12190 | -0.478 | 0.1730 | No | ||
| 45 | TOM1 | 13482 | -0.743 | 0.1141 | No | ||
| 46 | SLC17A5 | 13772 | -0.813 | 0.1101 | No | ||
| 47 | HPS1 | 14001 | -0.868 | 0.1102 | No | ||
| 48 | HYAL2 | 15028 | -1.143 | 0.0712 | No | ||
| 49 | ANKRD27 | 15214 | -1.193 | 0.0782 | No | ||
| 50 | ARSB | 15535 | -1.298 | 0.0794 | No | ||
| 51 | ABCA2 | 15657 | -1.340 | 0.0920 | No | ||
| 52 | TSPAN8 | 16509 | -1.682 | 0.0701 | No | ||
| 53 | PPT1 | 18057 | -3.044 | 0.0301 | No |