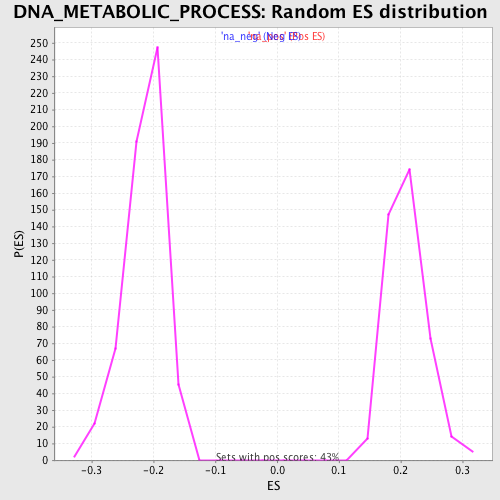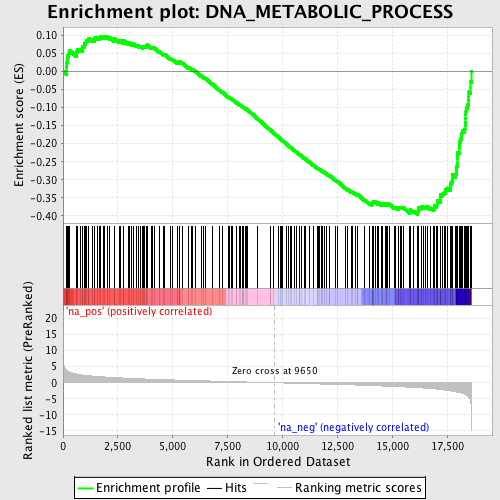
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | DNA_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.3958903 |
| Normalized Enrichment Score (NES) | -1.851916 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.058564242 |
| FWER p-Value | 0.656 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MSH5 | 154 | 3.949 | 0.0027 | No | ||
| 2 | LIG4 | 157 | 3.931 | 0.0137 | No | ||
| 3 | ATF7IP | 172 | 3.836 | 0.0238 | No | ||
| 4 | XRCC4 | 183 | 3.781 | 0.0339 | No | ||
| 5 | TFAM | 205 | 3.716 | 0.0432 | No | ||
| 6 | RAG1 | 266 | 3.461 | 0.0497 | No | ||
| 7 | UBE2A | 291 | 3.364 | 0.0578 | No | ||
| 8 | SMARCAD1 | 599 | 2.728 | 0.0488 | No | ||
| 9 | TERF2IP | 615 | 2.705 | 0.0556 | No | ||
| 10 | CDKN2A | 661 | 2.646 | 0.0606 | No | ||
| 11 | SUMO1 | 775 | 2.511 | 0.0616 | No | ||
| 12 | KPNA1 | 873 | 2.402 | 0.0630 | No | ||
| 13 | CDKN2D | 886 | 2.387 | 0.0691 | No | ||
| 14 | ORC5L | 957 | 2.331 | 0.0719 | No | ||
| 15 | RPA3 | 961 | 2.327 | 0.0783 | No | ||
| 16 | CIB1 | 1035 | 2.268 | 0.0807 | No | ||
| 17 | ERCC8 | 1068 | 2.247 | 0.0853 | No | ||
| 18 | KPNA2 | 1136 | 2.200 | 0.0878 | No | ||
| 19 | S100A11 | 1172 | 2.177 | 0.0921 | No | ||
| 20 | UBE2N | 1355 | 2.064 | 0.0880 | No | ||
| 21 | PTGES3 | 1432 | 2.013 | 0.0895 | No | ||
| 22 | TINF2 | 1452 | 1.995 | 0.0941 | No | ||
| 23 | IGF1 | 1567 | 1.934 | 0.0933 | No | ||
| 24 | CYCS | 1680 | 1.874 | 0.0925 | No | ||
| 25 | MSH6 | 1681 | 1.874 | 0.0978 | No | ||
| 26 | DDB1 | 1819 | 1.814 | 0.0955 | No | ||
| 27 | REPIN1 | 1908 | 1.767 | 0.0957 | No | ||
| 28 | ASF1A | 2000 | 1.725 | 0.0956 | No | ||
| 29 | TIPIN | 2131 | 1.671 | 0.0932 | No | ||
| 30 | CIDEA | 2325 | 1.604 | 0.0872 | No | ||
| 31 | ERCC4 | 2329 | 1.602 | 0.0916 | No | ||
| 32 | ATXN3 | 2565 | 1.526 | 0.0831 | No | ||
| 33 | POLD4 | 2599 | 1.516 | 0.0856 | No | ||
| 34 | RAD21 | 2736 | 1.470 | 0.0823 | No | ||
| 35 | RAD51L1 | 2761 | 1.459 | 0.0851 | No | ||
| 36 | PNKP | 2959 | 1.394 | 0.0783 | No | ||
| 37 | RAD17 | 3013 | 1.378 | 0.0793 | No | ||
| 38 | HMGB2 | 3135 | 1.340 | 0.0765 | No | ||
| 39 | XRN2 | 3203 | 1.316 | 0.0766 | No | ||
| 40 | TBRG1 | 3355 | 1.273 | 0.0720 | No | ||
| 41 | SMARCAL1 | 3437 | 1.246 | 0.0711 | No | ||
| 42 | GMNN | 3548 | 1.209 | 0.0685 | No | ||
| 43 | ATM | 3638 | 1.186 | 0.0670 | No | ||
| 44 | GADD45G | 3647 | 1.183 | 0.0699 | No | ||
| 45 | ADRA1D | 3725 | 1.162 | 0.0690 | No | ||
| 46 | MNAT1 | 3798 | 1.143 | 0.0683 | No | ||
| 47 | UBE2B | 3808 | 1.138 | 0.0710 | No | ||
| 48 | MSH3 | 3824 | 1.135 | 0.0734 | No | ||
| 49 | DNMT3A | 4030 | 1.088 | 0.0653 | No | ||
| 50 | FANCC | 4074 | 1.077 | 0.0660 | No | ||
| 51 | IGFBP4 | 4146 | 1.059 | 0.0651 | No | ||
| 52 | MYST2 | 4392 | 1.005 | 0.0546 | No | ||
| 53 | XRCC5 | 4569 | 0.965 | 0.0478 | No | ||
| 54 | MCM3AP | 4637 | 0.950 | 0.0468 | No | ||
| 55 | RFC4 | 4891 | 0.894 | 0.0356 | No | ||
| 56 | MUTYH | 4986 | 0.875 | 0.0329 | No | ||
| 57 | SMUG1 | 5191 | 0.835 | 0.0242 | No | ||
| 58 | TPD52L1 | 5206 | 0.832 | 0.0258 | No | ||
| 59 | GADD45A | 5285 | 0.815 | 0.0238 | No | ||
| 60 | XRCC6 | 5288 | 0.814 | 0.0260 | No | ||
| 61 | HMGB1 | 5302 | 0.811 | 0.0276 | No | ||
| 62 | APTX | 5422 | 0.784 | 0.0233 | No | ||
| 63 | MCM3 | 5720 | 0.721 | 0.0092 | No | ||
| 64 | KCTD13 | 5733 | 0.717 | 0.0106 | No | ||
| 65 | BTG2 | 5861 | 0.692 | 0.0056 | No | ||
| 66 | FEN1 | 5919 | 0.680 | 0.0044 | No | ||
| 67 | POLB | 6021 | 0.661 | 0.0008 | No | ||
| 68 | BNIP3 | 6288 | 0.613 | -0.0119 | No | ||
| 69 | TREX2 | 6406 | 0.590 | -0.0166 | No | ||
| 70 | SOD1 | 6486 | 0.576 | -0.0193 | No | ||
| 71 | CHEK1 | 6785 | 0.522 | -0.0340 | No | ||
| 72 | ORC3L | 7143 | 0.446 | -0.0522 | No | ||
| 73 | RAN | 7264 | 0.422 | -0.0575 | No | ||
| 74 | XRCC2 | 7529 | 0.380 | -0.0708 | No | ||
| 75 | POLA1 | 7588 | 0.368 | -0.0729 | No | ||
| 76 | MPG | 7665 | 0.353 | -0.0760 | No | ||
| 77 | RAD23B | 7714 | 0.344 | -0.0777 | No | ||
| 78 | RUVBL2 | 7879 | 0.315 | -0.0857 | No | ||
| 79 | ALKBH1 | 7924 | 0.307 | -0.0872 | No | ||
| 80 | DNASE2 | 8031 | 0.284 | -0.0922 | No | ||
| 81 | POT1 | 8070 | 0.279 | -0.0935 | No | ||
| 82 | RAD1 | 8160 | 0.265 | -0.0976 | No | ||
| 83 | GLI2 | 8173 | 0.262 | -0.0975 | No | ||
| 84 | TERF2 | 8216 | 0.255 | -0.0991 | No | ||
| 85 | ORC4L | 8236 | 0.251 | -0.0994 | No | ||
| 86 | FANCA | 8327 | 0.235 | -0.1036 | No | ||
| 87 | HUS1 | 8335 | 0.234 | -0.1033 | No | ||
| 88 | RFC3 | 8392 | 0.226 | -0.1057 | No | ||
| 89 | PMS1 | 8867 | 0.140 | -0.1311 | No | ||
| 90 | PPARGC1A | 9438 | 0.044 | -0.1620 | No | ||
| 91 | NF2 | 9447 | 0.043 | -0.1623 | No | ||
| 92 | PMS2 | 9581 | 0.014 | -0.1695 | No | ||
| 93 | TERT | 9808 | -0.031 | -0.1817 | No | ||
| 94 | ACHE | 9912 | -0.052 | -0.1871 | No | ||
| 95 | SETMAR | 9963 | -0.060 | -0.1897 | No | ||
| 96 | GTPBP4 | 10001 | -0.067 | -0.1915 | No | ||
| 97 | CD40LG | 10165 | -0.096 | -0.2001 | No | ||
| 98 | HTATIP | 10286 | -0.118 | -0.2063 | No | ||
| 99 | IL4 | 10370 | -0.134 | -0.2104 | No | ||
| 100 | TNP1 | 10389 | -0.138 | -0.2110 | No | ||
| 101 | MCM7 | 10535 | -0.166 | -0.2184 | No | ||
| 102 | ISG20 | 10622 | -0.179 | -0.2226 | No | ||
| 103 | IGHMBP2 | 10770 | -0.205 | -0.2300 | No | ||
| 104 | ATR | 10794 | -0.209 | -0.2307 | No | ||
| 105 | NUP98 | 10868 | -0.224 | -0.2340 | No | ||
| 106 | MCM2 | 11019 | -0.249 | -0.2415 | No | ||
| 107 | RNASEH2A | 11053 | -0.255 | -0.2426 | No | ||
| 108 | TLK2 | 11212 | -0.291 | -0.2503 | No | ||
| 109 | XRCC3 | 11389 | -0.323 | -0.2590 | No | ||
| 110 | POLH | 11578 | -0.358 | -0.2682 | No | ||
| 111 | PRIM1 | 11609 | -0.365 | -0.2688 | No | ||
| 112 | XAB2 | 11617 | -0.367 | -0.2681 | No | ||
| 113 | CECR2 | 11696 | -0.386 | -0.2713 | No | ||
| 114 | REV3L | 11777 | -0.400 | -0.2745 | No | ||
| 115 | AIFM1 | 11842 | -0.414 | -0.2768 | No | ||
| 116 | SYCP1 | 11925 | -0.429 | -0.2801 | No | ||
| 117 | POLE2 | 11991 | -0.438 | -0.2824 | No | ||
| 118 | TOP2A | 12133 | -0.467 | -0.2887 | No | ||
| 119 | REV1 | 12154 | -0.471 | -0.2885 | No | ||
| 120 | ORC2L | 12416 | -0.522 | -0.3012 | No | ||
| 121 | SMG6 | 12507 | -0.540 | -0.3046 | No | ||
| 122 | RAD51C | 12872 | -0.613 | -0.3227 | No | ||
| 123 | CDC6 | 12983 | -0.636 | -0.3268 | No | ||
| 124 | EGF | 13134 | -0.666 | -0.3331 | No | ||
| 125 | POLG | 13179 | -0.677 | -0.3336 | No | ||
| 126 | TGFB1 | 13308 | -0.704 | -0.3386 | No | ||
| 127 | GLI1 | 13401 | -0.726 | -0.3415 | No | ||
| 128 | UBE2V2 | 13435 | -0.732 | -0.3413 | No | ||
| 129 | EXO1 | 13747 | -0.808 | -0.3559 | No | ||
| 130 | HELLS | 13962 | -0.857 | -0.3651 | No | ||
| 131 | TNFSF13 | 14078 | -0.889 | -0.3689 | No | ||
| 132 | MSH2 | 14083 | -0.890 | -0.3666 | No | ||
| 133 | RBMS1 | 14088 | -0.891 | -0.3643 | No | ||
| 134 | SUPV3L1 | 14104 | -0.893 | -0.3626 | No | ||
| 135 | ERCC3 | 14137 | -0.903 | -0.3618 | No | ||
| 136 | TSN | 14159 | -0.909 | -0.3604 | No | ||
| 137 | ATRX | 14216 | -0.923 | -0.3608 | No | ||
| 138 | WRNIP1 | 14346 | -0.959 | -0.3651 | No | ||
| 139 | MGMT | 14355 | -0.963 | -0.3629 | No | ||
| 140 | POLD2 | 14522 | -1.009 | -0.3690 | No | ||
| 141 | SET | 14538 | -1.013 | -0.3670 | No | ||
| 142 | FOXL2 | 14558 | -1.018 | -0.3652 | No | ||
| 143 | MSH4 | 14673 | -1.045 | -0.3684 | No | ||
| 144 | ORC1L | 14735 | -1.063 | -0.3687 | No | ||
| 145 | GTF2H4 | 14739 | -1.064 | -0.3659 | No | ||
| 146 | DMC1 | 14768 | -1.072 | -0.3644 | No | ||
| 147 | RAD23A | 14873 | -1.100 | -0.3670 | No | ||
| 148 | RECQL | 15123 | -1.171 | -0.3772 | No | ||
| 149 | RAD51L3 | 15159 | -1.181 | -0.3758 | No | ||
| 150 | FANCG | 15268 | -1.211 | -0.3783 | No | ||
| 151 | DFFB | 15298 | -1.219 | -0.3764 | No | ||
| 152 | POLL | 15358 | -1.239 | -0.3761 | No | ||
| 153 | DNMT3B | 15431 | -1.268 | -0.3765 | No | ||
| 154 | PURA | 15510 | -1.289 | -0.3771 | No | ||
| 155 | CDC45L | 15791 | -1.386 | -0.3884 | No | ||
| 156 | OGG1 | 15818 | -1.398 | -0.3859 | No | ||
| 157 | BRCA1 | 15837 | -1.405 | -0.3829 | No | ||
| 158 | RAD52 | 15985 | -1.464 | -0.3868 | No | ||
| 159 | NTHL1 | 16154 | -1.533 | -0.3916 | Yes | ||
| 160 | NAP1L1 | 16172 | -1.539 | -0.3882 | Yes | ||
| 161 | CSNK1D | 16188 | -1.547 | -0.3846 | Yes | ||
| 162 | POLG2 | 16189 | -1.547 | -0.3803 | Yes | ||
| 163 | UNG | 16214 | -1.554 | -0.3772 | Yes | ||
| 164 | CDK2 | 16312 | -1.588 | -0.3780 | Yes | ||
| 165 | SMARCB1 | 16313 | -1.589 | -0.3735 | Yes | ||
| 166 | BRCA2 | 16426 | -1.645 | -0.3750 | Yes | ||
| 167 | POLQ | 16523 | -1.692 | -0.3754 | Yes | ||
| 168 | TDG | 16589 | -1.718 | -0.3741 | Yes | ||
| 169 | ERCC1 | 16754 | -1.814 | -0.3779 | Yes | ||
| 170 | RAD51 | 16891 | -1.889 | -0.3800 | Yes | ||
| 171 | DFFA | 16914 | -1.903 | -0.3758 | Yes | ||
| 172 | TEP1 | 16937 | -1.917 | -0.3716 | Yes | ||
| 173 | KIN | 17024 | -1.968 | -0.3708 | Yes | ||
| 174 | POLE3 | 17042 | -1.992 | -0.3661 | Yes | ||
| 175 | POLE | 17055 | -2.001 | -0.3611 | Yes | ||
| 176 | FOS | 17081 | -2.016 | -0.3568 | Yes | ||
| 177 | RPA1 | 17200 | -2.124 | -0.3572 | Yes | ||
| 178 | BLM | 17210 | -2.133 | -0.3517 | Yes | ||
| 179 | MRE11A | 17211 | -2.133 | -0.3457 | Yes | ||
| 180 | EREG | 17216 | -2.137 | -0.3399 | Yes | ||
| 181 | RECQL5 | 17311 | -2.213 | -0.3388 | Yes | ||
| 182 | NT5M | 17365 | -2.259 | -0.3353 | Yes | ||
| 183 | LIG1 | 17428 | -2.306 | -0.3322 | Yes | ||
| 184 | NASP | 17437 | -2.319 | -0.3261 | Yes | ||
| 185 | RAD9A | 17496 | -2.372 | -0.3225 | Yes | ||
| 186 | UPF1 | 17640 | -2.521 | -0.3232 | Yes | ||
| 187 | LIG3 | 17653 | -2.540 | -0.3167 | Yes | ||
| 188 | ERCC2 | 17660 | -2.555 | -0.3099 | Yes | ||
| 189 | DDB2 | 17719 | -2.609 | -0.3057 | Yes | ||
| 190 | CDK2AP1 | 17730 | -2.620 | -0.2988 | Yes | ||
| 191 | GTF2H1 | 17736 | -2.627 | -0.2917 | Yes | ||
| 192 | APEX1 | 17738 | -2.629 | -0.2844 | Yes | ||
| 193 | DNMT1 | 17892 | -2.816 | -0.2847 | Yes | ||
| 194 | MCM5 | 17916 | -2.853 | -0.2780 | Yes | ||
| 195 | RAD50 | 17930 | -2.875 | -0.2706 | Yes | ||
| 196 | POLI | 17938 | -2.884 | -0.2628 | Yes | ||
| 197 | IL7R | 17965 | -2.931 | -0.2560 | Yes | ||
| 198 | TLK1 | 17972 | -2.941 | -0.2480 | Yes | ||
| 199 | NBN | 17973 | -2.942 | -0.2397 | Yes | ||
| 200 | POLK | 17979 | -2.950 | -0.2317 | Yes | ||
| 201 | MLH1 | 17987 | -2.959 | -0.2238 | Yes | ||
| 202 | CDT1 | 18056 | -3.044 | -0.2189 | Yes | ||
| 203 | PPT1 | 18057 | -3.044 | -0.2103 | Yes | ||
| 204 | SPO11 | 18067 | -3.063 | -0.2022 | Yes | ||
| 205 | ABL1 | 18084 | -3.088 | -0.1943 | Yes | ||
| 206 | NT5E | 18123 | -3.158 | -0.1875 | Yes | ||
| 207 | XPC | 18146 | -3.203 | -0.1797 | Yes | ||
| 208 | UBE2V1 | 18169 | -3.244 | -0.1717 | Yes | ||
| 209 | DMAP1 | 18182 | -3.268 | -0.1632 | Yes | ||
| 210 | ERCC5 | 18295 | -3.572 | -0.1592 | Yes | ||
| 211 | RFC1 | 18319 | -3.654 | -0.1502 | Yes | ||
| 212 | VCP | 18328 | -3.709 | -0.1402 | Yes | ||
| 213 | RECQL4 | 18340 | -3.739 | -0.1302 | Yes | ||
| 214 | SMC1A | 18351 | -3.769 | -0.1202 | Yes | ||
| 215 | PICK1 | 18358 | -3.793 | -0.1098 | Yes | ||
| 216 | PARP1 | 18383 | -3.892 | -0.1002 | Yes | ||
| 217 | RAD54L | 18448 | -4.193 | -0.0918 | Yes | ||
| 218 | CSNK1E | 18481 | -4.377 | -0.0812 | Yes | ||
| 219 | NP | 18491 | -4.440 | -0.0692 | Yes | ||
| 220 | RPA2 | 18496 | -4.505 | -0.0568 | Yes | ||
| 221 | BAX | 18573 | -5.849 | -0.0444 | Yes | ||
| 222 | POLD1 | 18575 | -5.904 | -0.0278 | Yes | ||
| 223 | TP53 | 18610 | -10.661 | 0.0003 | Yes |