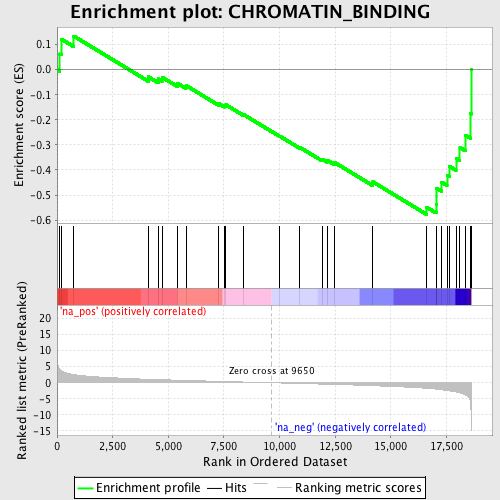
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHROMATIN_BINDING |
| Enrichment Score (ES) | -0.577405 |
| Normalized Enrichment Score (NES) | -1.9178364 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05067628 |
| FWER p-Value | 0.405 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL6 | 124 | 4.169 | 0.0626 | No | ||
| 2 | ING2 | 185 | 3.780 | 0.1222 | No | ||
| 3 | PAM | 751 | 2.532 | 0.1339 | No | ||
| 4 | MPO | 4100 | 1.072 | -0.0284 | No | ||
| 5 | NSD1 | 4556 | 0.967 | -0.0369 | No | ||
| 6 | EZH1 | 4717 | 0.932 | -0.0300 | No | ||
| 7 | APTX | 5422 | 0.784 | -0.0548 | No | ||
| 8 | RCC1 | 5794 | 0.704 | -0.0631 | No | ||
| 9 | RAN | 7264 | 0.422 | -0.1351 | No | ||
| 10 | FOXC2 | 7522 | 0.381 | -0.1426 | No | ||
| 11 | POLA1 | 7588 | 0.368 | -0.1400 | No | ||
| 12 | CDCA5 | 8391 | 0.226 | -0.1794 | No | ||
| 13 | KLHDC3 | 9996 | -0.067 | -0.2646 | No | ||
| 14 | SUV39H1 | 10915 | -0.232 | -0.3101 | No | ||
| 15 | TOP2B | 11908 | -0.425 | -0.3564 | No | ||
| 16 | TOP2A | 12133 | -0.467 | -0.3607 | No | ||
| 17 | CBX1 | 12479 | -0.536 | -0.3704 | No | ||
| 18 | CENPA | 14195 | -0.918 | -0.4474 | No | ||
| 19 | NCOA6 | 16613 | -1.736 | -0.5486 | Yes | ||
| 20 | CHAF1B | 17038 | -1.988 | -0.5383 | Yes | ||
| 21 | POLE | 17055 | -2.001 | -0.5059 | Yes | ||
| 22 | ACTL6A | 17056 | -2.001 | -0.4727 | Yes | ||
| 23 | CBX3 | 17294 | -2.194 | -0.4490 | Yes | ||
| 24 | CHAF1A | 17527 | -2.403 | -0.4215 | Yes | ||
| 25 | UPF1 | 17640 | -2.521 | -0.3857 | Yes | ||
| 26 | TOP1 | 17932 | -2.879 | -0.3535 | Yes | ||
| 27 | SMARCE1 | 18108 | -3.139 | -0.3107 | Yes | ||
| 28 | SMC1A | 18351 | -3.769 | -0.2611 | Yes | ||
| 29 | POLD1 | 18575 | -5.904 | -0.1750 | Yes | ||
| 30 | TP53 | 18610 | -10.661 | 0.0003 | Yes |