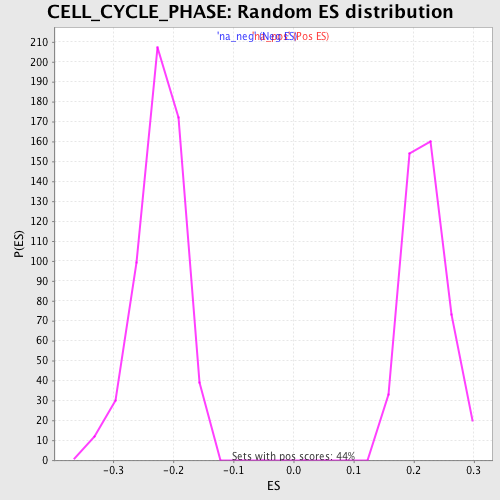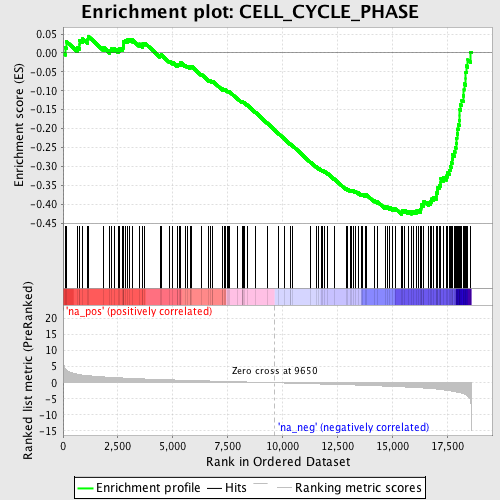
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CELL_CYCLE_PHASE |
| Enrichment Score (ES) | -0.42724714 |
| Normalized Enrichment Score (NES) | -1.9165748 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.047130026 |
| FWER p-Value | 0.412 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDKN1C | 103 | 4.297 | 0.0144 | No | ||
| 2 | MSH5 | 154 | 3.949 | 0.0300 | No | ||
| 3 | CDKN2A | 661 | 2.646 | 0.0149 | No | ||
| 4 | PAM | 751 | 2.532 | 0.0219 | No | ||
| 5 | CHMP1A | 754 | 2.528 | 0.0335 | No | ||
| 6 | CDKN2D | 886 | 2.387 | 0.0375 | No | ||
| 7 | MAD2L1 | 1128 | 2.202 | 0.0347 | No | ||
| 8 | KPNA2 | 1136 | 2.200 | 0.0446 | No | ||
| 9 | CCNA2 | 1853 | 1.799 | 0.0142 | No | ||
| 10 | TIPIN | 2131 | 1.671 | 0.0069 | No | ||
| 11 | CDKN1A | 2200 | 1.649 | 0.0109 | No | ||
| 12 | NPM2 | 2336 | 1.598 | 0.0110 | No | ||
| 13 | NUSAP1 | 2509 | 1.543 | 0.0089 | No | ||
| 14 | PIN1 | 2583 | 1.520 | 0.0120 | No | ||
| 15 | PPP6C | 2701 | 1.485 | 0.0126 | No | ||
| 16 | RAD21 | 2736 | 1.470 | 0.0176 | No | ||
| 17 | TTN | 2754 | 1.461 | 0.0234 | No | ||
| 18 | RAD51L1 | 2761 | 1.459 | 0.0299 | No | ||
| 19 | CUL4A | 2827 | 1.441 | 0.0331 | No | ||
| 20 | LATS2 | 2920 | 1.410 | 0.0346 | No | ||
| 21 | RAD17 | 3013 | 1.378 | 0.0361 | No | ||
| 22 | SUGT1 | 3148 | 1.333 | 0.0350 | No | ||
| 23 | DUSP13 | 3462 | 1.238 | 0.0238 | No | ||
| 24 | CDKN3 | 3626 | 1.190 | 0.0205 | No | ||
| 25 | ATM | 3638 | 1.186 | 0.0254 | No | ||
| 26 | CUL5 | 3722 | 1.163 | 0.0263 | No | ||
| 27 | ZWINT | 4415 | 1.000 | -0.0065 | No | ||
| 28 | ANAPC10 | 4467 | 0.987 | -0.0046 | No | ||
| 29 | TGFA | 4867 | 0.900 | -0.0221 | No | ||
| 30 | KATNA1 | 5004 | 0.872 | -0.0254 | No | ||
| 31 | TPD52L1 | 5206 | 0.832 | -0.0324 | No | ||
| 32 | NDC80 | 5232 | 0.826 | -0.0299 | No | ||
| 33 | CHFR | 5301 | 0.811 | -0.0298 | No | ||
| 34 | CDC23 | 5353 | 0.800 | -0.0288 | No | ||
| 35 | NEK2 | 5362 | 0.796 | -0.0256 | No | ||
| 36 | PIM2 | 5562 | 0.756 | -0.0328 | No | ||
| 37 | ANAPC11 | 5680 | 0.730 | -0.0358 | No | ||
| 38 | APBB2 | 5787 | 0.706 | -0.0382 | No | ||
| 39 | RCC1 | 5794 | 0.704 | -0.0353 | No | ||
| 40 | HSPA2 | 5849 | 0.695 | -0.0350 | No | ||
| 41 | ANLN | 6293 | 0.612 | -0.0561 | No | ||
| 42 | PTPRC | 6637 | 0.548 | -0.0721 | No | ||
| 43 | USH1C | 6713 | 0.535 | -0.0737 | No | ||
| 44 | CHEK1 | 6785 | 0.522 | -0.0751 | No | ||
| 45 | CDKN2B | 6825 | 0.514 | -0.0749 | No | ||
| 46 | RAN | 7264 | 0.422 | -0.0966 | No | ||
| 47 | UBE2C | 7270 | 0.421 | -0.0949 | No | ||
| 48 | MAD2L2 | 7364 | 0.408 | -0.0981 | No | ||
| 49 | BOLL | 7388 | 0.403 | -0.0974 | No | ||
| 50 | CDKN1B | 7498 | 0.383 | -0.1016 | No | ||
| 51 | XRCC2 | 7529 | 0.380 | -0.1014 | No | ||
| 52 | POLA1 | 7588 | 0.368 | -0.1028 | No | ||
| 53 | RB1 | 7963 | 0.297 | -0.1217 | No | ||
| 54 | PRMT5 | 8157 | 0.265 | -0.1309 | No | ||
| 55 | RAD1 | 8160 | 0.265 | -0.1298 | No | ||
| 56 | ANAPC4 | 8162 | 0.265 | -0.1286 | No | ||
| 57 | BUB1 | 8225 | 0.254 | -0.1308 | No | ||
| 58 | CCNA1 | 8247 | 0.249 | -0.1308 | No | ||
| 59 | CDCA5 | 8391 | 0.226 | -0.1375 | No | ||
| 60 | TIMELESS | 8756 | 0.160 | -0.1564 | No | ||
| 61 | AURKA | 9293 | 0.067 | -0.1851 | No | ||
| 62 | CUL2 | 9838 | -0.039 | -0.2144 | No | ||
| 63 | STAG3 | 10080 | -0.083 | -0.2271 | No | ||
| 64 | NEK6 | 10344 | -0.127 | -0.2407 | No | ||
| 65 | CENPE | 10373 | -0.134 | -0.2416 | No | ||
| 66 | CDC16 | 10470 | -0.152 | -0.2461 | No | ||
| 67 | CD28 | 11286 | -0.304 | -0.2888 | No | ||
| 68 | CETN1 | 11531 | -0.350 | -0.3004 | No | ||
| 69 | KIF11 | 11659 | -0.378 | -0.3055 | No | ||
| 70 | BUB1B | 11770 | -0.400 | -0.3096 | No | ||
| 71 | CUL3 | 11837 | -0.412 | -0.3113 | No | ||
| 72 | SYCP1 | 11925 | -0.429 | -0.3140 | No | ||
| 73 | TOP3A | 12047 | -0.450 | -0.3184 | No | ||
| 74 | BIRC5 | 12364 | -0.512 | -0.3332 | No | ||
| 75 | EPGN | 12897 | -0.619 | -0.3591 | No | ||
| 76 | CDC6 | 12983 | -0.636 | -0.3607 | No | ||
| 77 | TTK | 13086 | -0.658 | -0.3632 | No | ||
| 78 | EGF | 13134 | -0.666 | -0.3627 | No | ||
| 79 | CDK6 | 13219 | -0.686 | -0.3640 | No | ||
| 80 | TGFB1 | 13308 | -0.704 | -0.3655 | No | ||
| 81 | SSSCA1 | 13452 | -0.737 | -0.3698 | No | ||
| 82 | AKAP8 | 13583 | -0.766 | -0.3733 | No | ||
| 83 | DCTN3 | 13654 | -0.783 | -0.3734 | No | ||
| 84 | KIF15 | 13789 | -0.817 | -0.3769 | No | ||
| 85 | SMC4 | 13804 | -0.821 | -0.3738 | No | ||
| 86 | GFI1B | 14183 | -0.915 | -0.3901 | No | ||
| 87 | CUL1 | 14329 | -0.955 | -0.3935 | No | ||
| 88 | MSH4 | 14673 | -1.045 | -0.4072 | No | ||
| 89 | DMC1 | 14768 | -1.072 | -0.4073 | No | ||
| 90 | CDC7 | 14883 | -1.103 | -0.4083 | No | ||
| 91 | ACVR1 | 15033 | -1.143 | -0.4111 | No | ||
| 92 | RAD51L3 | 15159 | -1.181 | -0.4124 | No | ||
| 93 | KNTC1 | 15435 | -1.269 | -0.4214 | Yes | ||
| 94 | KHDRBS1 | 15450 | -1.270 | -0.4162 | Yes | ||
| 95 | BCAT1 | 15569 | -1.312 | -0.4165 | Yes | ||
| 96 | SMC3 | 15722 | -1.364 | -0.4184 | Yes | ||
| 97 | ZW10 | 15866 | -1.415 | -0.4196 | Yes | ||
| 98 | RAD52 | 15985 | -1.464 | -0.4191 | Yes | ||
| 99 | E2F1 | 16090 | -1.502 | -0.4178 | Yes | ||
| 100 | CDK4 | 16191 | -1.548 | -0.4160 | Yes | ||
| 101 | TARDBP | 16295 | -1.582 | -0.4142 | Yes | ||
| 102 | CDK2 | 16312 | -1.588 | -0.4077 | Yes | ||
| 103 | MAP3K11 | 16319 | -1.593 | -0.4006 | Yes | ||
| 104 | CENPF | 16425 | -1.644 | -0.3987 | Yes | ||
| 105 | CDC25B | 16441 | -1.655 | -0.3918 | Yes | ||
| 106 | NCAPH | 16656 | -1.758 | -0.3952 | Yes | ||
| 107 | PCBP4 | 16759 | -1.815 | -0.3923 | Yes | ||
| 108 | GSPT1 | 16804 | -1.840 | -0.3862 | Yes | ||
| 109 | RAD51 | 16891 | -1.889 | -0.3820 | Yes | ||
| 110 | TBRG4 | 17003 | -1.957 | -0.3789 | Yes | ||
| 111 | CDC25C | 17009 | -1.959 | -0.3701 | Yes | ||
| 112 | POLE | 17055 | -2.001 | -0.3632 | Yes | ||
| 113 | SKP2 | 17079 | -2.016 | -0.3551 | Yes | ||
| 114 | PPP5C | 17161 | -2.096 | -0.3498 | Yes | ||
| 115 | MRE11A | 17211 | -2.133 | -0.3425 | Yes | ||
| 116 | EREG | 17216 | -2.137 | -0.3328 | Yes | ||
| 117 | CIT | 17333 | -2.227 | -0.3287 | Yes | ||
| 118 | TPX2 | 17473 | -2.353 | -0.3253 | Yes | ||
| 119 | GFI1 | 17529 | -2.405 | -0.3171 | Yes | ||
| 120 | ANAPC5 | 17619 | -2.499 | -0.3103 | Yes | ||
| 121 | LIG3 | 17653 | -2.540 | -0.3003 | Yes | ||
| 122 | KIF22 | 17690 | -2.585 | -0.2903 | Yes | ||
| 123 | CDK2AP1 | 17730 | -2.620 | -0.2802 | Yes | ||
| 124 | NOLC1 | 17748 | -2.639 | -0.2688 | Yes | ||
| 125 | PLK1 | 17847 | -2.764 | -0.2613 | Yes | ||
| 126 | NUMA1 | 17864 | -2.791 | -0.2492 | Yes | ||
| 127 | DDX11 | 17909 | -2.844 | -0.2384 | Yes | ||
| 128 | RAD50 | 17930 | -2.875 | -0.2261 | Yes | ||
| 129 | CLIP1 | 17953 | -2.913 | -0.2137 | Yes | ||
| 130 | NBN | 17973 | -2.942 | -0.2011 | Yes | ||
| 131 | CDK10 | 18008 | -2.992 | -0.1890 | Yes | ||
| 132 | SPO11 | 18067 | -3.063 | -0.1779 | Yes | ||
| 133 | PML | 18083 | -3.084 | -0.1644 | Yes | ||
| 134 | ABL1 | 18084 | -3.088 | -0.1501 | Yes | ||
| 135 | MPHOSPH6 | 18116 | -3.151 | -0.1371 | Yes | ||
| 136 | ACVR1B | 18173 | -3.255 | -0.1250 | Yes | ||
| 137 | DCTN2 | 18256 | -3.458 | -0.1134 | Yes | ||
| 138 | DLG7 | 18270 | -3.498 | -0.0978 | Yes | ||
| 139 | CDKN2C | 18274 | -3.503 | -0.0817 | Yes | ||
| 140 | KIF2C | 18349 | -3.764 | -0.0682 | Yes | ||
| 141 | SMC1A | 18351 | -3.769 | -0.0508 | Yes | ||
| 142 | APBB1 | 18379 | -3.882 | -0.0342 | Yes | ||
| 143 | RAD54L | 18448 | -4.193 | -0.0184 | Yes | ||
| 144 | POLD1 | 18575 | -5.904 | 0.0022 | Yes |