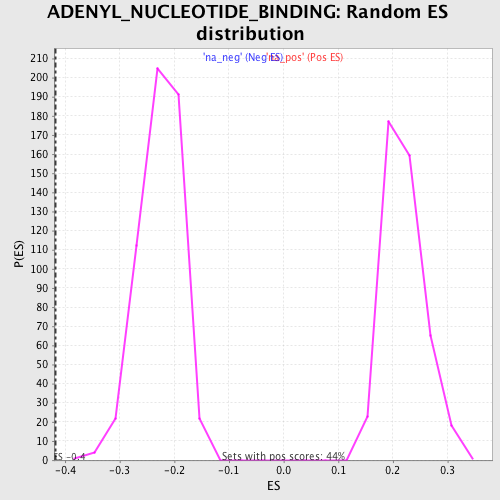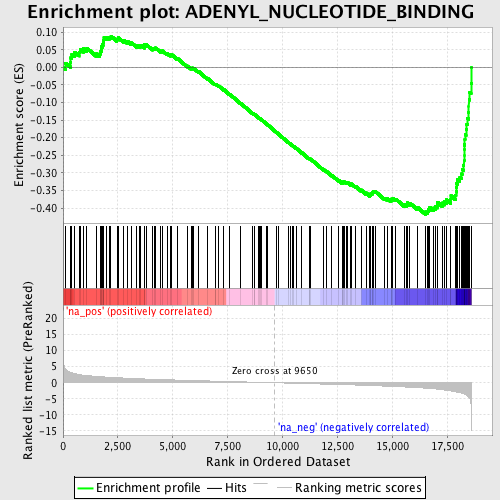
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ADENYL_NUCLEOTIDE_BINDING |
| Enrichment Score (ES) | -0.41834307 |
| Normalized Enrichment Score (NES) | -1.8519232 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.062009197 |
| FWER p-Value | 0.656 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PINK1 | 109 | 4.262 | 0.0124 | No | ||
| 2 | STK17B | 341 | 3.238 | 0.0138 | No | ||
| 3 | TGFBR1 | 354 | 3.193 | 0.0268 | No | ||
| 4 | DDX25 | 400 | 3.076 | 0.0376 | No | ||
| 5 | SNRK | 517 | 2.872 | 0.0436 | No | ||
| 6 | RPS6KA4 | 726 | 2.562 | 0.0433 | No | ||
| 7 | ATP7A | 771 | 2.513 | 0.0517 | No | ||
| 8 | ABCC3 | 926 | 2.355 | 0.0535 | No | ||
| 9 | TNK1 | 1081 | 2.233 | 0.0547 | No | ||
| 10 | ABCB7 | 1536 | 1.951 | 0.0385 | No | ||
| 11 | MSH6 | 1681 | 1.874 | 0.0387 | No | ||
| 12 | AIFM2 | 1701 | 1.866 | 0.0457 | No | ||
| 13 | BCKDK | 1738 | 1.850 | 0.0517 | No | ||
| 14 | TOR1A | 1749 | 1.846 | 0.0591 | No | ||
| 15 | MAP4K5 | 1780 | 1.832 | 0.0653 | No | ||
| 16 | PGK1 | 1822 | 1.812 | 0.0709 | No | ||
| 17 | PPP2R4 | 1846 | 1.802 | 0.0773 | No | ||
| 18 | ABCG1 | 1862 | 1.795 | 0.0842 | No | ||
| 19 | IRAK3 | 1977 | 1.735 | 0.0855 | No | ||
| 20 | ABCD2 | 2097 | 1.685 | 0.0863 | No | ||
| 21 | TWF2 | 2181 | 1.655 | 0.0889 | No | ||
| 22 | MAST1 | 2456 | 1.561 | 0.0808 | No | ||
| 23 | WNK3 | 2511 | 1.543 | 0.0844 | No | ||
| 24 | TTN | 2754 | 1.461 | 0.0776 | No | ||
| 25 | LATS2 | 2920 | 1.410 | 0.0747 | No | ||
| 26 | MAP3K5 | 3121 | 1.343 | 0.0697 | No | ||
| 27 | BMPR1A | 3362 | 1.271 | 0.0621 | No | ||
| 28 | MTRR | 3491 | 1.229 | 0.0605 | No | ||
| 29 | STK4 | 3535 | 1.212 | 0.0633 | No | ||
| 30 | SMARCA5 | 3693 | 1.170 | 0.0598 | No | ||
| 31 | GABARAPL2 | 3696 | 1.169 | 0.0648 | No | ||
| 32 | WNK4 | 3796 | 1.144 | 0.0643 | No | ||
| 33 | CDKL5 | 4075 | 1.076 | 0.0539 | No | ||
| 34 | ACVRL1 | 4147 | 1.059 | 0.0546 | No | ||
| 35 | SNF1LK2 | 4217 | 1.044 | 0.0553 | No | ||
| 36 | MYO6 | 4460 | 0.988 | 0.0464 | No | ||
| 37 | GALK1 | 4529 | 0.971 | 0.0469 | No | ||
| 38 | RPS6KA5 | 4740 | 0.928 | 0.0395 | No | ||
| 39 | BRSK2 | 4879 | 0.898 | 0.0359 | No | ||
| 40 | NLRC4 | 4925 | 0.885 | 0.0373 | No | ||
| 41 | ABCB6 | 5196 | 0.834 | 0.0262 | No | ||
| 42 | STK38 | 5667 | 0.733 | 0.0039 | No | ||
| 43 | CLP1 | 5834 | 0.697 | -0.0021 | No | ||
| 44 | MAP4K3 | 5882 | 0.689 | -0.0016 | No | ||
| 45 | EIF2B2 | 5940 | 0.676 | -0.0018 | No | ||
| 46 | ERN2 | 6185 | 0.631 | -0.0123 | No | ||
| 47 | DGKE | 6580 | 0.561 | -0.0313 | No | ||
| 48 | ABCC6 | 6923 | 0.492 | -0.0477 | No | ||
| 49 | DAPK2 | 6956 | 0.484 | -0.0473 | No | ||
| 50 | PRKCI | 7099 | 0.456 | -0.0530 | No | ||
| 51 | TSSK6 | 7325 | 0.414 | -0.0634 | No | ||
| 52 | CDC42BPB | 7600 | 0.365 | -0.0767 | No | ||
| 53 | MAP4K1 | 8082 | 0.277 | -0.1016 | No | ||
| 54 | TRNT1 | 8633 | 0.184 | -0.1305 | No | ||
| 55 | NEK11 | 8717 | 0.167 | -0.1343 | No | ||
| 56 | APRT | 8732 | 0.164 | -0.1344 | No | ||
| 57 | EIF4A3 | 8909 | 0.133 | -0.1433 | No | ||
| 58 | SEPHS1 | 8940 | 0.127 | -0.1444 | No | ||
| 59 | GCK | 8991 | 0.118 | -0.1466 | No | ||
| 60 | GCLC | 9027 | 0.112 | -0.1480 | No | ||
| 61 | PXK | 9262 | 0.074 | -0.1604 | No | ||
| 62 | PRKACB | 9310 | 0.065 | -0.1626 | No | ||
| 63 | MOV10L1 | 9705 | -0.011 | -0.1839 | No | ||
| 64 | VPS4A | 9836 | -0.039 | -0.1908 | No | ||
| 65 | TSSK3 | 10257 | -0.112 | -0.2131 | No | ||
| 66 | NEK6 | 10344 | -0.127 | -0.2172 | No | ||
| 67 | ABCA4 | 10451 | -0.149 | -0.2223 | No | ||
| 68 | CFTR | 10513 | -0.161 | -0.2249 | No | ||
| 69 | ABCC1 | 10637 | -0.181 | -0.2308 | No | ||
| 70 | CDC42BPA | 10883 | -0.226 | -0.2430 | No | ||
| 71 | TLK2 | 11212 | -0.291 | -0.2596 | No | ||
| 72 | MAP4K4 | 11215 | -0.292 | -0.2584 | No | ||
| 73 | NLRP1 | 11294 | -0.305 | -0.2613 | No | ||
| 74 | ERN1 | 11871 | -0.418 | -0.2907 | No | ||
| 75 | TRIB1 | 11999 | -0.440 | -0.2957 | No | ||
| 76 | BTK | 12246 | -0.490 | -0.3069 | No | ||
| 77 | SNF1LK | 12554 | -0.549 | -0.3212 | No | ||
| 78 | BMPR1B | 12733 | -0.587 | -0.3283 | No | ||
| 79 | HSP90AA1 | 12753 | -0.590 | -0.3268 | No | ||
| 80 | ABCB11 | 12778 | -0.595 | -0.3255 | No | ||
| 81 | MARK1 | 12829 | -0.605 | -0.3257 | No | ||
| 82 | GSG2 | 12936 | -0.627 | -0.3287 | No | ||
| 83 | HSPA4L | 12953 | -0.630 | -0.3269 | No | ||
| 84 | ABCC8 | 13107 | -0.661 | -0.3323 | No | ||
| 85 | WNK1 | 13142 | -0.668 | -0.3313 | No | ||
| 86 | TSSK2 | 13331 | -0.710 | -0.3384 | No | ||
| 87 | N4BP2 | 13581 | -0.766 | -0.3486 | No | ||
| 88 | SMC4 | 13804 | -0.821 | -0.3571 | No | ||
| 89 | ABCG2 | 13980 | -0.861 | -0.3629 | No | ||
| 90 | DAPK3 | 14027 | -0.875 | -0.3616 | No | ||
| 91 | ABCA3 | 14029 | -0.875 | -0.3579 | No | ||
| 92 | MSH2 | 14083 | -0.890 | -0.3570 | No | ||
| 93 | PIM1 | 14109 | -0.895 | -0.3545 | No | ||
| 94 | MKNK2 | 14143 | -0.904 | -0.3524 | No | ||
| 95 | STK3 | 14230 | -0.926 | -0.3531 | No | ||
| 96 | PPOX | 14665 | -1.044 | -0.3721 | No | ||
| 97 | DMC1 | 14768 | -1.072 | -0.3730 | No | ||
| 98 | DGKZ | 14947 | -1.121 | -0.3779 | No | ||
| 99 | NVL | 14971 | -1.127 | -0.3743 | No | ||
| 100 | ACVR1 | 15033 | -1.143 | -0.3727 | No | ||
| 101 | MARK2 | 15162 | -1.181 | -0.3745 | No | ||
| 102 | TSSK1A | 15571 | -1.313 | -0.3910 | No | ||
| 103 | ABCA2 | 15657 | -1.340 | -0.3898 | No | ||
| 104 | NDOR1 | 15678 | -1.346 | -0.3852 | No | ||
| 105 | NLK | 15807 | -1.393 | -0.3861 | No | ||
| 106 | STK38L | 16139 | -1.522 | -0.3975 | No | ||
| 107 | ATP7B | 16525 | -1.693 | -0.4111 | Yes | ||
| 108 | DAPK1 | 16625 | -1.742 | -0.4090 | Yes | ||
| 109 | ABCA7 | 16661 | -1.760 | -0.4033 | Yes | ||
| 110 | SARS2 | 16713 | -1.791 | -0.3984 | Yes | ||
| 111 | RAD51 | 16891 | -1.889 | -0.3999 | Yes | ||
| 112 | NOX4 | 16976 | -1.939 | -0.3961 | Yes | ||
| 113 | MINK1 | 17048 | -1.995 | -0.3914 | Yes | ||
| 114 | MYO3A | 17062 | -2.005 | -0.3835 | Yes | ||
| 115 | PGK2 | 17275 | -2.182 | -0.3856 | Yes | ||
| 116 | ICK | 17383 | -2.270 | -0.3817 | Yes | ||
| 117 | TPX2 | 17473 | -2.353 | -0.3764 | Yes | ||
| 118 | MYH9 | 17671 | -2.566 | -0.3761 | Yes | ||
| 119 | MAP3K6 | 17678 | -2.574 | -0.3654 | Yes | ||
| 120 | ACTA1 | 17879 | -2.807 | -0.3641 | Yes | ||
| 121 | MAP4K2 | 17907 | -2.832 | -0.3535 | Yes | ||
| 122 | NME1 | 17927 | -2.871 | -0.3422 | Yes | ||
| 123 | ABCF2 | 17941 | -2.887 | -0.3305 | Yes | ||
| 124 | TLK1 | 17972 | -2.941 | -0.3195 | Yes | ||
| 125 | ABL1 | 18084 | -3.088 | -0.3123 | Yes | ||
| 126 | ACVR1B | 18173 | -3.255 | -0.3031 | Yes | ||
| 127 | PEX6 | 18185 | -3.270 | -0.2897 | Yes | ||
| 128 | MAST2 | 18242 | -3.430 | -0.2780 | Yes | ||
| 129 | STK11 | 18271 | -3.499 | -0.2645 | Yes | ||
| 130 | DYRK3 | 18277 | -3.516 | -0.2497 | Yes | ||
| 131 | ABCF1 | 18280 | -3.530 | -0.2347 | Yes | ||
| 132 | ABCC5 | 18285 | -3.539 | -0.2197 | Yes | ||
| 133 | MYO9B | 18314 | -3.639 | -0.2056 | Yes | ||
| 134 | RFC1 | 18319 | -3.654 | -0.1902 | Yes | ||
| 135 | MKNK1 | 18365 | -3.823 | -0.1762 | Yes | ||
| 136 | CDC2L2 | 18400 | -3.964 | -0.1610 | Yes | ||
| 137 | SPHK1 | 18431 | -4.103 | -0.1451 | Yes | ||
| 138 | SRPK1 | 18488 | -4.410 | -0.1292 | Yes | ||
| 139 | ABCD4 | 18497 | -4.508 | -0.1103 | Yes | ||
| 140 | DHX38 | 18499 | -4.522 | -0.0909 | Yes | ||
| 141 | SRPK2 | 18537 | -4.940 | -0.0718 | Yes | ||
| 142 | MYH10 | 18593 | -7.046 | -0.0445 | Yes | ||
| 143 | TP53 | 18610 | -10.661 | 0.0003 | Yes |