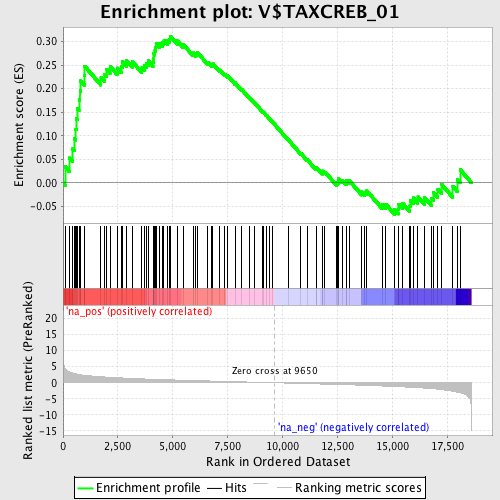
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$TAXCREB_01 |
| Enrichment Score (ES) | 0.3115778 |
| Normalized Enrichment Score (NES) | 1.3109531 |
| Nominal p-value | 0.06609808 |
| FDR q-value | 0.91691107 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MESDC1 | 93 | 4.458 | 0.0343 | Yes | ||
| 2 | CYLD | 296 | 3.352 | 0.0530 | Yes | ||
| 3 | CALM2 | 433 | 3.005 | 0.0722 | Yes | ||
| 4 | CDC42 | 507 | 2.885 | 0.0937 | Yes | ||
| 5 | SDHB | 581 | 2.756 | 0.1141 | Yes | ||
| 6 | NINJ1 | 604 | 2.716 | 0.1369 | Yes | ||
| 7 | MBNL2 | 647 | 2.659 | 0.1581 | Yes | ||
| 8 | ARIH1 | 728 | 2.561 | 0.1763 | Yes | ||
| 9 | PLK4 | 769 | 2.513 | 0.1964 | Yes | ||
| 10 | SMARCD1 | 803 | 2.488 | 0.2165 | Yes | ||
| 11 | PAFAH1B1 | 972 | 2.320 | 0.2279 | Yes | ||
| 12 | LUC7L2 | 987 | 2.305 | 0.2475 | Yes | ||
| 13 | POLDIP3 | 1725 | 1.857 | 0.2241 | Yes | ||
| 14 | APPBP2 | 1902 | 1.769 | 0.2302 | Yes | ||
| 15 | ATF1 | 1985 | 1.732 | 0.2411 | Yes | ||
| 16 | SLC30A5 | 2141 | 1.667 | 0.2474 | Yes | ||
| 17 | DLAT | 2462 | 1.557 | 0.2439 | Yes | ||
| 18 | PAK1 | 2648 | 1.501 | 0.2472 | Yes | ||
| 19 | SLC25A5 | 2694 | 1.486 | 0.2579 | Yes | ||
| 20 | GTF2A1 | 2882 | 1.422 | 0.2603 | Yes | ||
| 21 | NR2F1 | 3152 | 1.332 | 0.2575 | Yes | ||
| 22 | XPR1 | 3593 | 1.196 | 0.2443 | Yes | ||
| 23 | SMARCA5 | 3693 | 1.170 | 0.2493 | Yes | ||
| 24 | DLX5 | 3793 | 1.145 | 0.2541 | Yes | ||
| 25 | RING1 | 3881 | 1.121 | 0.2593 | Yes | ||
| 26 | CAMK2D | 4099 | 1.072 | 0.2570 | Yes | ||
| 27 | USP14 | 4125 | 1.064 | 0.2651 | Yes | ||
| 28 | KLF13 | 4131 | 1.062 | 0.2742 | Yes | ||
| 29 | NR4A3 | 4185 | 1.050 | 0.2806 | Yes | ||
| 30 | PTPRU | 4215 | 1.045 | 0.2882 | Yes | ||
| 31 | OSR1 | 4236 | 1.041 | 0.2963 | Yes | ||
| 32 | RAB37 | 4398 | 1.003 | 0.2965 | Yes | ||
| 33 | OTUB2 | 4539 | 0.969 | 0.2975 | Yes | ||
| 34 | SHANK2 | 4597 | 0.960 | 0.3029 | Yes | ||
| 35 | HOXA10 | 4762 | 0.921 | 0.3022 | Yes | ||
| 36 | NOL4 | 4851 | 0.905 | 0.3054 | Yes | ||
| 37 | TAF11 | 4884 | 0.895 | 0.3116 | Yes | ||
| 38 | GEM | 5204 | 0.833 | 0.3017 | No | ||
| 39 | PEG3 | 5466 | 0.775 | 0.2945 | No | ||
| 40 | PPP2CB | 5920 | 0.679 | 0.2760 | No | ||
| 41 | HAS1 | 6053 | 0.654 | 0.2746 | No | ||
| 42 | PNRC1 | 6125 | 0.641 | 0.2765 | No | ||
| 43 | SFRS1 | 6599 | 0.557 | 0.2558 | No | ||
| 44 | RFX5 | 6761 | 0.527 | 0.2518 | No | ||
| 45 | CEBPB | 6830 | 0.513 | 0.2527 | No | ||
| 46 | SSTR3 | 7141 | 0.446 | 0.2399 | No | ||
| 47 | GPR158 | 7371 | 0.407 | 0.2311 | No | ||
| 48 | CDKN1B | 7498 | 0.383 | 0.2277 | No | ||
| 49 | OSBP | 7836 | 0.323 | 0.2123 | No | ||
| 50 | DLST | 8133 | 0.270 | 0.1987 | No | ||
| 51 | CDK5R2 | 8509 | 0.206 | 0.1803 | No | ||
| 52 | JMJD1B | 8712 | 0.167 | 0.1709 | No | ||
| 53 | CD2AP | 9093 | 0.101 | 0.1513 | No | ||
| 54 | LMO4 | 9122 | 0.096 | 0.1506 | No | ||
| 55 | SOX10 | 9259 | 0.074 | 0.1439 | No | ||
| 56 | GPM6B | 9424 | 0.046 | 0.1355 | No | ||
| 57 | HHIP | 9548 | 0.023 | 0.1290 | No | ||
| 58 | MLF2 | 10267 | -0.114 | 0.0913 | No | ||
| 59 | HS6ST2 | 10836 | -0.218 | 0.0625 | No | ||
| 60 | SIX3 | 11122 | -0.270 | 0.0495 | No | ||
| 61 | MAFF | 11526 | -0.348 | 0.0308 | No | ||
| 62 | SCAMP5 | 11550 | -0.354 | 0.0327 | No | ||
| 63 | PHF12 | 11812 | -0.408 | 0.0222 | No | ||
| 64 | ARRB2 | 11822 | -0.410 | 0.0253 | No | ||
| 65 | KDELR3 | 11934 | -0.430 | 0.0232 | No | ||
| 66 | GPR3 | 12471 | -0.534 | -0.0011 | No | ||
| 67 | FGF14 | 12514 | -0.542 | 0.0014 | No | ||
| 68 | SNF1LK | 12554 | -0.549 | 0.0042 | No | ||
| 69 | MAP1LC3A | 12564 | -0.551 | 0.0086 | No | ||
| 70 | BNIP2 | 12724 | -0.585 | 0.0051 | No | ||
| 71 | G6PC | 12900 | -0.620 | 0.0012 | No | ||
| 72 | CLSTN3 | 12924 | -0.623 | 0.0054 | No | ||
| 73 | FOSB | 13036 | -0.648 | 0.0051 | No | ||
| 74 | MARCKS | 13610 | -0.772 | -0.0190 | No | ||
| 75 | PCF11 | 13753 | -0.810 | -0.0195 | No | ||
| 76 | NTRK2 | 13843 | -0.829 | -0.0170 | No | ||
| 77 | EIF4A2 | 14548 | -1.017 | -0.0460 | No | ||
| 78 | RPS27 | 14700 | -1.052 | -0.0449 | No | ||
| 79 | IRX4 | 15112 | -1.167 | -0.0568 | No | ||
| 80 | TAGLN2 | 15285 | -1.216 | -0.0553 | No | ||
| 81 | CTCF | 15305 | -1.221 | -0.0456 | No | ||
| 82 | GPC6 | 15482 | -1.280 | -0.0438 | No | ||
| 83 | NLK | 15807 | -1.393 | -0.0490 | No | ||
| 84 | HOXB1 | 15829 | -1.401 | -0.0377 | No | ||
| 85 | DUSP1 | 15957 | -1.454 | -0.0318 | No | ||
| 86 | ARMCX3 | 16174 | -1.540 | -0.0298 | No | ||
| 87 | GATA1 | 16481 | -1.670 | -0.0316 | No | ||
| 88 | GSPT1 | 16804 | -1.840 | -0.0328 | No | ||
| 89 | PHF16 | 16879 | -1.879 | -0.0202 | No | ||
| 90 | FOS | 17081 | -2.016 | -0.0132 | No | ||
| 91 | CCND2 | 17244 | -2.160 | -0.0029 | No | ||
| 92 | TRIM39 | 17765 | -2.663 | -0.0075 | No | ||
| 93 | VAMP2 | 17961 | -2.926 | 0.0078 | No | ||
| 94 | PLCB3 | 18096 | -3.117 | 0.0281 | No |
