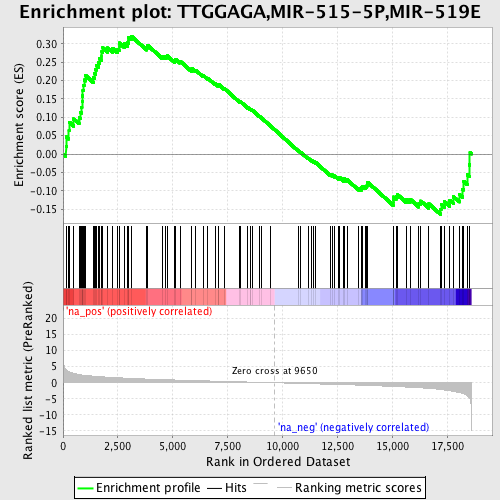
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | 0.32104665 |
| Normalized Enrichment Score (NES) | 1.3627076 |
| Nominal p-value | 0.03275109 |
| FDR q-value | 0.92958945 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAP2C | 131 | 4.135 | 0.0210 | Yes | ||
| 2 | SERTAD1 | 152 | 3.974 | 0.0469 | Yes | ||
| 3 | CMTM6 | 267 | 3.461 | 0.0642 | Yes | ||
| 4 | RAP1B | 276 | 3.413 | 0.0869 | Yes | ||
| 5 | YTHDF2 | 483 | 2.926 | 0.0956 | Yes | ||
| 6 | ZBTB1 | 731 | 2.557 | 0.0996 | Yes | ||
| 7 | CLCF1 | 792 | 2.494 | 0.1133 | Yes | ||
| 8 | SGPL1 | 829 | 2.452 | 0.1280 | Yes | ||
| 9 | RPP14 | 861 | 2.415 | 0.1427 | Yes | ||
| 10 | KPNA1 | 873 | 2.402 | 0.1584 | Yes | ||
| 11 | DCX | 902 | 2.378 | 0.1730 | Yes | ||
| 12 | ABCC3 | 926 | 2.355 | 0.1878 | Yes | ||
| 13 | ZFP36L1 | 960 | 2.328 | 0.2018 | Yes | ||
| 14 | SP4 | 1017 | 2.281 | 0.2142 | Yes | ||
| 15 | RSBN1 | 1381 | 2.043 | 0.2085 | Yes | ||
| 16 | CLSTN2 | 1438 | 2.009 | 0.2191 | Yes | ||
| 17 | CAB39 | 1479 | 1.983 | 0.2304 | Yes | ||
| 18 | HNRPUL1 | 1514 | 1.963 | 0.2419 | Yes | ||
| 19 | YAF2 | 1615 | 1.911 | 0.2494 | Yes | ||
| 20 | SUMO2 | 1669 | 1.881 | 0.2593 | Yes | ||
| 21 | HOXA1 | 1760 | 1.840 | 0.2669 | Yes | ||
| 22 | GABBR1 | 1766 | 1.836 | 0.2791 | Yes | ||
| 23 | EPC2 | 1810 | 1.819 | 0.2892 | Yes | ||
| 24 | RDH11 | 2029 | 1.711 | 0.2890 | Yes | ||
| 25 | AP3S1 | 2247 | 1.632 | 0.2883 | Yes | ||
| 26 | NUP35 | 2490 | 1.548 | 0.2858 | Yes | ||
| 27 | APLN | 2558 | 1.528 | 0.2925 | Yes | ||
| 28 | ATXN3 | 2565 | 1.526 | 0.3025 | Yes | ||
| 29 | SOX30 | 2790 | 1.452 | 0.3003 | Yes | ||
| 30 | POU2F1 | 2923 | 1.410 | 0.3027 | Yes | ||
| 31 | RIOK3 | 2977 | 1.390 | 0.3093 | Yes | ||
| 32 | ANUBL1 | 2982 | 1.387 | 0.3185 | Yes | ||
| 33 | KCND2 | 3105 | 1.348 | 0.3210 | Yes | ||
| 34 | UBE2B | 3808 | 1.138 | 0.2909 | No | ||
| 35 | OPHN1 | 3855 | 1.127 | 0.2960 | No | ||
| 36 | CSNK1G1 | 4527 | 0.972 | 0.2664 | No | ||
| 37 | DDX3Y | 4665 | 0.942 | 0.2654 | No | ||
| 38 | TGFBR2 | 4739 | 0.928 | 0.2677 | No | ||
| 39 | DDX3X | 5080 | 0.857 | 0.2552 | No | ||
| 40 | ARRDC3 | 5125 | 0.848 | 0.2585 | No | ||
| 41 | DGCR2 | 5327 | 0.806 | 0.2531 | No | ||
| 42 | BTG2 | 5861 | 0.692 | 0.2290 | No | ||
| 43 | ETNK2 | 5868 | 0.691 | 0.2334 | No | ||
| 44 | SYNGR1 | 6037 | 0.657 | 0.2288 | No | ||
| 45 | UBE2H | 6378 | 0.595 | 0.2145 | No | ||
| 46 | SFRS1 | 6599 | 0.557 | 0.2064 | No | ||
| 47 | PLAGL2 | 6931 | 0.490 | 0.1918 | No | ||
| 48 | PDGFRA | 7065 | 0.464 | 0.1878 | No | ||
| 49 | ACY1L2 | 7071 | 0.463 | 0.1906 | No | ||
| 50 | MOSPD2 | 7362 | 0.408 | 0.1777 | No | ||
| 51 | ATRN | 8043 | 0.283 | 0.1429 | No | ||
| 52 | EPN2 | 8094 | 0.276 | 0.1421 | No | ||
| 53 | NFIB | 8421 | 0.222 | 0.1260 | No | ||
| 54 | POM121 | 8518 | 0.205 | 0.1222 | No | ||
| 55 | RPS6KA3 | 8560 | 0.198 | 0.1213 | No | ||
| 56 | FSTL1 | 8624 | 0.186 | 0.1192 | No | ||
| 57 | ARHGEF3 | 8962 | 0.124 | 0.1018 | No | ||
| 58 | ISL1 | 9028 | 0.112 | 0.0991 | No | ||
| 59 | SLC9A2 | 9429 | 0.046 | 0.0778 | No | ||
| 60 | MLLT6 | 10714 | -0.197 | 0.0098 | No | ||
| 61 | CPEB2 | 10827 | -0.216 | 0.0052 | No | ||
| 62 | SNAP25 | 11173 | -0.282 | -0.0115 | No | ||
| 63 | SLC7A11 | 11316 | -0.309 | -0.0171 | No | ||
| 64 | TNPO1 | 11421 | -0.329 | -0.0205 | No | ||
| 65 | IQGAP1 | 11525 | -0.348 | -0.0237 | No | ||
| 66 | PUM1 | 12170 | -0.473 | -0.0552 | No | ||
| 67 | NOTCH2 | 12262 | -0.493 | -0.0568 | No | ||
| 68 | KCTD7 | 12376 | -0.514 | -0.0594 | No | ||
| 69 | NFATC4 | 12565 | -0.551 | -0.0659 | No | ||
| 70 | BSN | 12587 | -0.557 | -0.0632 | No | ||
| 71 | FREQ | 12798 | -0.598 | -0.0705 | No | ||
| 72 | NCOA1 | 12805 | -0.599 | -0.0668 | No | ||
| 73 | MTHFD1L | 12944 | -0.628 | -0.0700 | No | ||
| 74 | PSMD11 | 13467 | -0.740 | -0.0931 | No | ||
| 75 | SCN8A | 13584 | -0.766 | -0.0942 | No | ||
| 76 | MARCKS | 13610 | -0.772 | -0.0903 | No | ||
| 77 | DONSON | 13657 | -0.784 | -0.0875 | No | ||
| 78 | EFNA3 | 13793 | -0.818 | -0.0892 | No | ||
| 79 | SLC4A4 | 13848 | -0.831 | -0.0865 | No | ||
| 80 | EMX2 | 13860 | -0.834 | -0.0814 | No | ||
| 81 | FRAS1 | 13884 | -0.838 | -0.0770 | No | ||
| 82 | ETS1 | 15043 | -1.147 | -0.1318 | No | ||
| 83 | MAPRE1 | 15047 | -1.148 | -0.1241 | No | ||
| 84 | ITSN1 | 15058 | -1.150 | -0.1169 | No | ||
| 85 | FGFR2 | 15176 | -1.184 | -0.1152 | No | ||
| 86 | RBL2 | 15239 | -1.201 | -0.1104 | No | ||
| 87 | TMEM47 | 15649 | -1.338 | -0.1234 | No | ||
| 88 | MEF2D | 15813 | -1.397 | -0.1227 | No | ||
| 89 | ULK2 | 16215 | -1.554 | -0.1338 | No | ||
| 90 | ARID1A | 16304 | -1.584 | -0.1278 | No | ||
| 91 | HIP2 | 16672 | -1.765 | -0.1357 | No | ||
| 92 | MTF2 | 17208 | -2.131 | -0.1501 | No | ||
| 93 | ETV1 | 17229 | -2.147 | -0.1366 | No | ||
| 94 | ICK | 17383 | -2.270 | -0.1295 | No | ||
| 95 | SOX12 | 17622 | -2.504 | -0.1254 | No | ||
| 96 | CYR61 | 17792 | -2.700 | -0.1162 | No | ||
| 97 | TMEM2 | 18060 | -3.049 | -0.1099 | No | ||
| 98 | GDI1 | 18209 | -3.336 | -0.0953 | No | ||
| 99 | ABCG4 | 18262 | -3.473 | -0.0745 | No | ||
| 100 | UBAP2 | 18418 | -4.015 | -0.0557 | No | ||
| 101 | OSBPL3 | 18516 | -4.714 | -0.0289 | No | ||
| 102 | UHRF2 | 18544 | -5.049 | 0.0039 | No |
