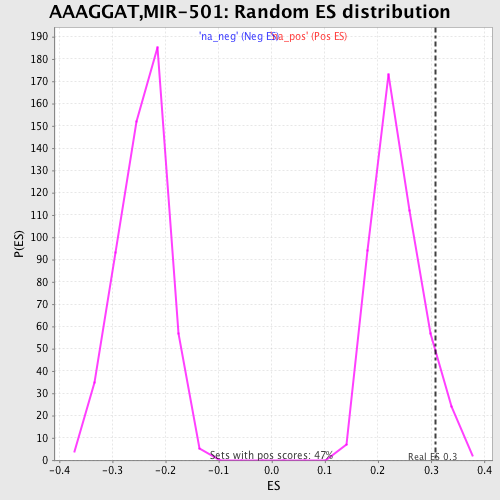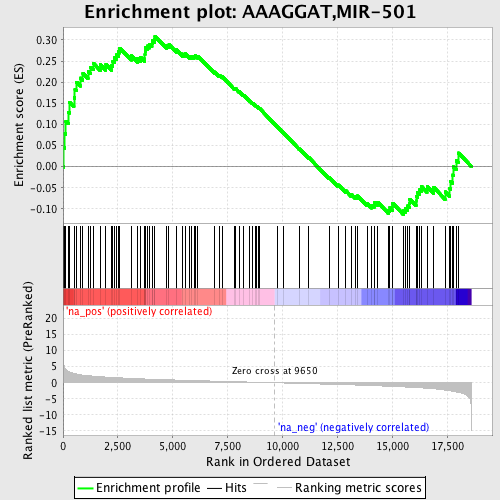
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | AAAGGAT,MIR-501 |
| Enrichment Score (ES) | 0.30907091 |
| Normalized Enrichment Score (NES) | 1.3089811 |
| Nominal p-value | 0.07249467 |
| FDR q-value | 0.8327048 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MYLK | 21 | 6.191 | 0.0462 | Yes | ||
| 2 | UBAP1 | 79 | 4.622 | 0.0785 | Yes | ||
| 3 | BCL6 | 124 | 4.169 | 0.1081 | Yes | ||
| 4 | ACADSB | 240 | 3.561 | 0.1291 | Yes | ||
| 5 | PIK3AP1 | 284 | 3.393 | 0.1527 | Yes | ||
| 6 | RNF11 | 497 | 2.900 | 0.1635 | Yes | ||
| 7 | CUGBP2 | 539 | 2.833 | 0.1829 | Yes | ||
| 8 | RABGEF1 | 605 | 2.714 | 0.2002 | Yes | ||
| 9 | SGPP1 | 781 | 2.507 | 0.2099 | Yes | ||
| 10 | DCX | 902 | 2.378 | 0.2217 | Yes | ||
| 11 | TAF5L | 1138 | 2.198 | 0.2258 | Yes | ||
| 12 | CD164 | 1255 | 2.125 | 0.2358 | Yes | ||
| 13 | RSBN1 | 1381 | 2.043 | 0.2447 | Yes | ||
| 14 | ADIPOR2 | 1698 | 1.866 | 0.2419 | Yes | ||
| 15 | HES5 | 1949 | 1.748 | 0.2418 | Yes | ||
| 16 | RAB22A | 2219 | 1.643 | 0.2398 | Yes | ||
| 17 | PPP1CB | 2266 | 1.625 | 0.2498 | Yes | ||
| 18 | H2AFX | 2344 | 1.595 | 0.2578 | Yes | ||
| 19 | ATP6V1H | 2439 | 1.566 | 0.2647 | Yes | ||
| 20 | TNNI2 | 2520 | 1.537 | 0.2722 | Yes | ||
| 21 | RGL1 | 2580 | 1.522 | 0.2806 | Yes | ||
| 22 | KCND2 | 3105 | 1.348 | 0.2626 | Yes | ||
| 23 | CART1 | 3393 | 1.259 | 0.2568 | Yes | ||
| 24 | SOX4 | 3534 | 1.213 | 0.2585 | Yes | ||
| 25 | SNAP29 | 3726 | 1.162 | 0.2571 | Yes | ||
| 26 | MEF2C | 3730 | 1.161 | 0.2658 | Yes | ||
| 27 | PHF6 | 3735 | 1.159 | 0.2744 | Yes | ||
| 28 | CTDSP1 | 3757 | 1.153 | 0.2821 | Yes | ||
| 29 | GIF | 3850 | 1.128 | 0.2858 | Yes | ||
| 30 | USP12 | 3942 | 1.105 | 0.2893 | Yes | ||
| 31 | LPIN1 | 4052 | 1.083 | 0.2917 | Yes | ||
| 32 | VDAC2 | 4067 | 1.079 | 0.2992 | Yes | ||
| 33 | ELAVL4 | 4175 | 1.052 | 0.3015 | Yes | ||
| 34 | NR4A3 | 4185 | 1.050 | 0.3091 | Yes | ||
| 35 | PITX2 | 4716 | 0.932 | 0.2876 | No | ||
| 36 | RDX | 4825 | 0.910 | 0.2887 | No | ||
| 37 | CUTL2 | 5159 | 0.842 | 0.2772 | No | ||
| 38 | SYT7 | 5462 | 0.776 | 0.2668 | No | ||
| 39 | KCNRG | 5557 | 0.757 | 0.2675 | No | ||
| 40 | MAP2K1 | 5762 | 0.712 | 0.2620 | No | ||
| 41 | CLK1 | 5869 | 0.691 | 0.2615 | No | ||
| 42 | TOMM70A | 5967 | 0.672 | 0.2614 | No | ||
| 43 | ROBO2 | 6013 | 0.663 | 0.2641 | No | ||
| 44 | PDK1 | 6145 | 0.638 | 0.2619 | No | ||
| 45 | CELSR2 | 6912 | 0.494 | 0.2243 | No | ||
| 46 | SCN3A | 7133 | 0.449 | 0.2159 | No | ||
| 47 | HAS2 | 7245 | 0.426 | 0.2131 | No | ||
| 48 | NFASC | 7803 | 0.329 | 0.1856 | No | ||
| 49 | SLC25A3 | 7838 | 0.323 | 0.1862 | No | ||
| 50 | NFIL3 | 8017 | 0.288 | 0.1788 | No | ||
| 51 | SATB2 | 8224 | 0.254 | 0.1696 | No | ||
| 52 | SLITRK5 | 8512 | 0.205 | 0.1557 | No | ||
| 53 | RET | 8641 | 0.183 | 0.1502 | No | ||
| 54 | LEPROTL1 | 8746 | 0.161 | 0.1458 | No | ||
| 55 | PRKCE | 8821 | 0.148 | 0.1430 | No | ||
| 56 | SYNC1 | 8890 | 0.136 | 0.1403 | No | ||
| 57 | SEPHS1 | 8940 | 0.127 | 0.1386 | No | ||
| 58 | AMMECR1 | 9754 | -0.021 | 0.0949 | No | ||
| 59 | CHODL | 10046 | -0.075 | 0.0798 | No | ||
| 60 | HOXB8 | 10772 | -0.205 | 0.0422 | No | ||
| 61 | SENP3 | 11201 | -0.289 | 0.0213 | No | ||
| 62 | NR2F2 | 12123 | -0.465 | -0.0249 | No | ||
| 63 | NPR3 | 12542 | -0.547 | -0.0432 | No | ||
| 64 | SFRS2 | 12880 | -0.616 | -0.0567 | No | ||
| 65 | SLC35B3 | 13150 | -0.670 | -0.0661 | No | ||
| 66 | QKI | 13327 | -0.709 | -0.0702 | No | ||
| 67 | GRB10 | 13411 | -0.728 | -0.0691 | No | ||
| 68 | WDFY3 | 13852 | -0.831 | -0.0865 | No | ||
| 69 | ZIC4 | 14066 | -0.886 | -0.0912 | No | ||
| 70 | LRRC1 | 14174 | -0.913 | -0.0900 | No | ||
| 71 | RCN1 | 14204 | -0.920 | -0.0845 | No | ||
| 72 | CUL1 | 14329 | -0.955 | -0.0839 | No | ||
| 73 | UBE2Q1 | 14843 | -1.093 | -0.1033 | No | ||
| 74 | KIF1C | 14894 | -1.105 | -0.0975 | No | ||
| 75 | SOX11 | 15015 | -1.140 | -0.0953 | No | ||
| 76 | PLXNB1 | 15020 | -1.141 | -0.0868 | No | ||
| 77 | PURA | 15510 | -1.289 | -0.1033 | No | ||
| 78 | UBE4B | 15598 | -1.323 | -0.0979 | No | ||
| 79 | MAP3K8 | 15699 | -1.352 | -0.0929 | No | ||
| 80 | ALS2 | 15780 | -1.381 | -0.0867 | No | ||
| 81 | PNN | 15800 | -1.389 | -0.0771 | No | ||
| 82 | PPP2R5E | 16088 | -1.501 | -0.0811 | No | ||
| 83 | NFIX | 16092 | -1.503 | -0.0697 | No | ||
| 84 | SRR | 16150 | -1.531 | -0.0611 | No | ||
| 85 | BCLAF1 | 16256 | -1.568 | -0.0548 | No | ||
| 86 | ADCYAP1 | 16327 | -1.595 | -0.0464 | No | ||
| 87 | DNAJB12 | 16587 | -1.717 | -0.0472 | No | ||
| 88 | PHF16 | 16879 | -1.879 | -0.0485 | No | ||
| 89 | MYB | 17411 | -2.294 | -0.0597 | No | ||
| 90 | PHC1 | 17620 | -2.499 | -0.0518 | No | ||
| 91 | ZC3H7A | 17654 | -2.541 | -0.0341 | No | ||
| 92 | TRIM39 | 17765 | -2.663 | -0.0197 | No | ||
| 93 | JUN | 17790 | -2.698 | -0.0003 | No | ||
| 94 | ACACA | 17919 | -2.859 | 0.0146 | No | ||
| 95 | CLK2 | 18015 | -2.997 | 0.0324 | No |