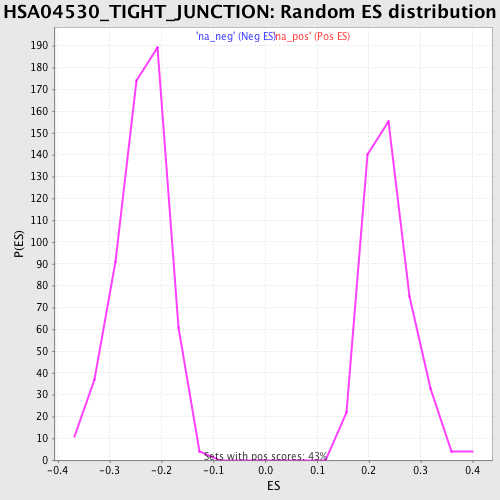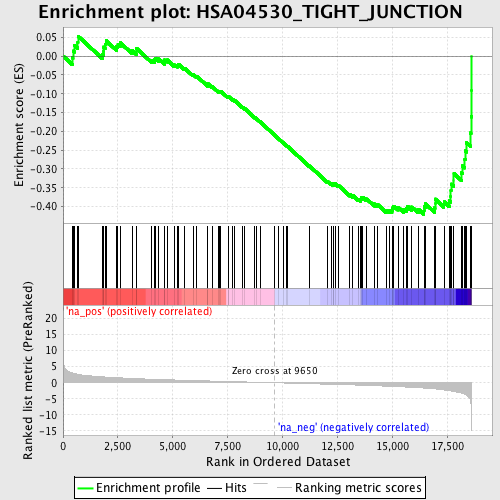
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04530_TIGHT_JUNCTION |
| Enrichment Score (ES) | -0.4210759 |
| Normalized Enrichment Score (NES) | -1.760086 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.33104956 |
| FWER p-Value | 0.722 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MYL2 | 425 | 3.024 | -0.0038 | No | ||
| 2 | RRAS | 451 | 2.973 | 0.0137 | No | ||
| 3 | CDC42 | 507 | 2.885 | 0.0291 | No | ||
| 4 | MYLPF | 672 | 2.637 | 0.0369 | No | ||
| 5 | MYL9 | 679 | 2.624 | 0.0532 | No | ||
| 6 | JAM2 | 1795 | 1.825 | 0.0046 | No | ||
| 7 | PPP2R4 | 1846 | 1.802 | 0.0133 | No | ||
| 8 | CLDN4 | 1854 | 1.799 | 0.0243 | No | ||
| 9 | PTEN | 1917 | 1.762 | 0.0322 | No | ||
| 10 | MYH11 | 1956 | 1.745 | 0.0412 | No | ||
| 11 | VAPA | 2441 | 1.565 | 0.0250 | No | ||
| 12 | ACTG1 | 2500 | 1.546 | 0.0316 | No | ||
| 13 | EXOC3 | 2600 | 1.516 | 0.0359 | No | ||
| 14 | PARD6A | 3149 | 1.333 | 0.0148 | No | ||
| 15 | CLDN1 | 3343 | 1.280 | 0.0124 | No | ||
| 16 | AKT3 | 3346 | 1.278 | 0.0204 | No | ||
| 17 | RAB13 | 4047 | 1.083 | -0.0105 | No | ||
| 18 | MYL7 | 4160 | 1.056 | -0.0098 | No | ||
| 19 | MYH2 | 4197 | 1.048 | -0.0051 | No | ||
| 20 | CSNK2A1 | 4357 | 1.014 | -0.0073 | No | ||
| 21 | CRB3 | 4630 | 0.952 | -0.0159 | No | ||
| 22 | PRKCZ | 4634 | 0.951 | -0.0101 | No | ||
| 23 | CLDN9 | 4742 | 0.926 | -0.0100 | No | ||
| 24 | CLDN23 | 5092 | 0.855 | -0.0234 | No | ||
| 25 | CLDN19 | 5199 | 0.833 | -0.0239 | No | ||
| 26 | RHOA | 5246 | 0.824 | -0.0211 | No | ||
| 27 | PARD6B | 5531 | 0.762 | -0.0316 | No | ||
| 28 | PPP2CB | 5920 | 0.679 | -0.0483 | No | ||
| 29 | F11R | 6097 | 0.647 | -0.0537 | No | ||
| 30 | JAM3 | 6595 | 0.557 | -0.0770 | No | ||
| 31 | PPP2R2A | 6596 | 0.557 | -0.0734 | No | ||
| 32 | PARD6G | 6787 | 0.521 | -0.0804 | No | ||
| 33 | PRKCI | 7099 | 0.456 | -0.0943 | No | ||
| 34 | CLDN5 | 7136 | 0.448 | -0.0934 | No | ||
| 35 | MPDZ | 7186 | 0.438 | -0.0933 | No | ||
| 36 | PRKCQ | 7521 | 0.381 | -0.1089 | No | ||
| 37 | CLDN11 | 7537 | 0.378 | -0.1073 | No | ||
| 38 | MRAS | 7722 | 0.343 | -0.1151 | No | ||
| 39 | PPP2R1A | 7811 | 0.328 | -0.1177 | No | ||
| 40 | SRC | 8170 | 0.263 | -0.1354 | No | ||
| 41 | HRAS | 8277 | 0.244 | -0.1396 | No | ||
| 42 | CLDN8 | 8743 | 0.162 | -0.1637 | No | ||
| 43 | PRKCE | 8821 | 0.148 | -0.1669 | No | ||
| 44 | MYH3 | 8978 | 0.121 | -0.1745 | No | ||
| 45 | MYH6 | 9651 | -0.000 | -0.2108 | No | ||
| 46 | NRAS | 9797 | -0.029 | -0.2185 | No | ||
| 47 | CSNK2B | 10022 | -0.070 | -0.2301 | No | ||
| 48 | GNAI1 | 10182 | -0.098 | -0.2381 | No | ||
| 49 | AKT1 | 10228 | -0.108 | -0.2398 | No | ||
| 50 | PPP2CA | 11227 | -0.293 | -0.2918 | No | ||
| 51 | RAB3B | 12039 | -0.449 | -0.3328 | No | ||
| 52 | KRAS | 12215 | -0.484 | -0.3392 | No | ||
| 53 | CLDN16 | 12312 | -0.503 | -0.3412 | No | ||
| 54 | ASH1L | 12333 | -0.507 | -0.3390 | No | ||
| 55 | MYH4 | 12401 | -0.518 | -0.3394 | No | ||
| 56 | CLDN2 | 12534 | -0.546 | -0.3430 | No | ||
| 57 | GNAI3 | 13072 | -0.655 | -0.3679 | No | ||
| 58 | PRKCB1 | 13209 | -0.683 | -0.3709 | No | ||
| 59 | CASK | 13474 | -0.741 | -0.3804 | No | ||
| 60 | ACTB | 13575 | -0.764 | -0.3810 | No | ||
| 61 | CLDN14 | 13582 | -0.766 | -0.3765 | No | ||
| 62 | RRAS2 | 13667 | -0.786 | -0.3760 | No | ||
| 63 | CLDN17 | 13834 | -0.827 | -0.3797 | No | ||
| 64 | CLDN18 | 14187 | -0.917 | -0.3929 | No | ||
| 65 | CLDN7 | 14327 | -0.953 | -0.3944 | No | ||
| 66 | MYH14 | 14743 | -1.065 | -0.4101 | No | ||
| 67 | CLDN15 | 14871 | -1.100 | -0.4099 | No | ||
| 68 | OCLN | 14990 | -1.132 | -0.4091 | No | ||
| 69 | PARD3 | 15024 | -1.142 | -0.4037 | No | ||
| 70 | YES1 | 15065 | -1.152 | -0.3985 | No | ||
| 71 | CGN | 15265 | -1.209 | -0.4016 | No | ||
| 72 | CTTN | 15525 | -1.293 | -0.4074 | No | ||
| 73 | TJP1 | 15639 | -1.335 | -0.4050 | No | ||
| 74 | AKT2 | 15689 | -1.350 | -0.3991 | No | ||
| 75 | ACTN2 | 15894 | -1.429 | -0.4011 | No | ||
| 76 | CDK4 | 16191 | -1.548 | -0.4073 | No | ||
| 77 | CLDN10 | 16448 | -1.657 | -0.4106 | Yes | ||
| 78 | MYH7 | 16454 | -1.658 | -0.4003 | Yes | ||
| 79 | PRKCA | 16510 | -1.682 | -0.3926 | Yes | ||
| 80 | ACTN3 | 16938 | -1.917 | -0.4035 | Yes | ||
| 81 | CLDN3 | 16951 | -1.928 | -0.3920 | Yes | ||
| 82 | PRKCD | 16955 | -1.929 | -0.3799 | Yes | ||
| 83 | CSDA | 17360 | -2.253 | -0.3874 | Yes | ||
| 84 | MPP5 | 17603 | -2.490 | -0.3847 | Yes | ||
| 85 | MYH9 | 17671 | -2.566 | -0.3720 | Yes | ||
| 86 | PRKCH | 17677 | -2.572 | -0.3560 | Yes | ||
| 87 | PPM1J | 17684 | -2.577 | -0.3400 | Yes | ||
| 88 | ACTN4 | 17813 | -2.730 | -0.3296 | Yes | ||
| 89 | MAGI1 | 17814 | -2.731 | -0.3123 | Yes | ||
| 90 | TJP2 | 18156 | -3.212 | -0.3103 | Yes | ||
| 91 | TJP3 | 18202 | -3.319 | -0.2917 | Yes | ||
| 92 | INADL | 18311 | -3.624 | -0.2746 | Yes | ||
| 93 | HCLS1 | 18326 | -3.704 | -0.2518 | Yes | ||
| 94 | CSNK2A2 | 18390 | -3.916 | -0.2304 | Yes | ||
| 95 | EXOC4 | 18563 | -5.605 | -0.2042 | Yes | ||
| 96 | MYH10 | 18593 | -7.046 | -0.1611 | Yes | ||
| 97 | MYLC2PL | 18611 | -10.977 | -0.0924 | Yes | ||
| 98 | AMOTL1 | 18615 | -14.604 | 0.0001 | Yes |