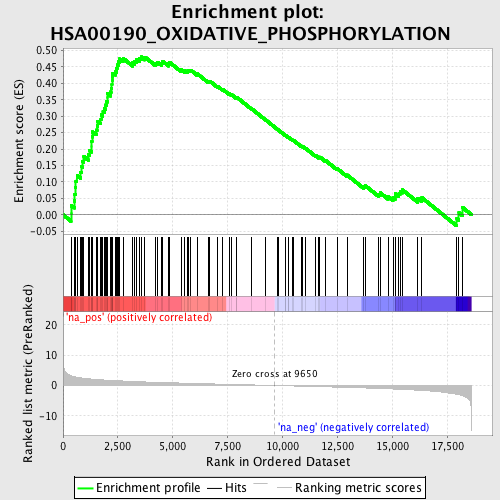
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA00190_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.48061758 |
| Normalized Enrichment Score (NES) | 2.0692694 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.024643809 |
| FWER p-Value | 0.025 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NDUFA6 | 361 | 3.163 | 0.0045 | Yes | ||
| 2 | ATP6V1A | 363 | 3.160 | 0.0285 | Yes | ||
| 3 | NDUFB3 | 512 | 2.877 | 0.0424 | Yes | ||
| 4 | COX7A2 | 531 | 2.844 | 0.0630 | Yes | ||
| 5 | UQCRH | 562 | 2.785 | 0.0826 | Yes | ||
| 6 | SDHB | 581 | 2.756 | 0.1026 | Yes | ||
| 7 | ATP6V1F | 632 | 2.684 | 0.1203 | Yes | ||
| 8 | NDUFA1 | 801 | 2.490 | 0.1301 | Yes | ||
| 9 | NDUFS4 | 831 | 2.449 | 0.1472 | Yes | ||
| 10 | NDUFB5 | 888 | 2.387 | 0.1623 | Yes | ||
| 11 | ATP6V0E1 | 946 | 2.339 | 0.1770 | Yes | ||
| 12 | ATP5F1 | 1148 | 2.190 | 0.1828 | Yes | ||
| 13 | ATP6V1B2 | 1195 | 2.157 | 0.1967 | Yes | ||
| 14 | ATP6AP1 | 1306 | 2.096 | 0.2067 | Yes | ||
| 15 | NDUFB9 | 1307 | 2.096 | 0.2226 | Yes | ||
| 16 | SDHC | 1320 | 2.086 | 0.2379 | Yes | ||
| 17 | NDUFB4 | 1344 | 2.072 | 0.2524 | Yes | ||
| 18 | NDUFA13 | 1515 | 1.963 | 0.2581 | Yes | ||
| 19 | NDUFS3 | 1549 | 1.946 | 0.2711 | Yes | ||
| 20 | UQCRFS1 | 1579 | 1.930 | 0.2842 | Yes | ||
| 21 | UQCRB | 1719 | 1.859 | 0.2909 | Yes | ||
| 22 | NDUFB8 | 1735 | 1.850 | 0.3041 | Yes | ||
| 23 | NDUFA11 | 1817 | 1.815 | 0.3135 | Yes | ||
| 24 | NDUFA5 | 1887 | 1.779 | 0.3233 | Yes | ||
| 25 | ATP5E | 1934 | 1.754 | 0.3342 | Yes | ||
| 26 | ATP12A | 1984 | 1.732 | 0.3447 | Yes | ||
| 27 | NDUFC2 | 2016 | 1.717 | 0.3561 | Yes | ||
| 28 | NDUFB2 | 2018 | 1.716 | 0.3691 | Yes | ||
| 29 | NDUFA2 | 2164 | 1.660 | 0.3739 | Yes | ||
| 30 | COX6C | 2186 | 1.653 | 0.3853 | Yes | ||
| 31 | ATP6V1D | 2225 | 1.641 | 0.3957 | Yes | ||
| 32 | SDHD | 2230 | 1.637 | 0.4079 | Yes | ||
| 33 | COX6B1 | 2239 | 1.635 | 0.4199 | Yes | ||
| 34 | NDUFA7 | 2256 | 1.629 | 0.4314 | Yes | ||
| 35 | COX7A1 | 2373 | 1.585 | 0.4372 | Yes | ||
| 36 | ATP6V1H | 2439 | 1.566 | 0.4456 | Yes | ||
| 37 | ATP6V0D1 | 2464 | 1.557 | 0.4562 | Yes | ||
| 38 | NDUFA9 | 2510 | 1.543 | 0.4655 | Yes | ||
| 39 | NDUFA4 | 2561 | 1.527 | 0.4744 | Yes | ||
| 40 | ATP5I | 2735 | 1.471 | 0.4762 | Yes | ||
| 41 | NDUFS8 | 3177 | 1.326 | 0.4625 | Yes | ||
| 42 | NDUFA3 | 3263 | 1.301 | 0.4678 | Yes | ||
| 43 | NDUFS2 | 3336 | 1.281 | 0.4736 | Yes | ||
| 44 | NDUFB10 | 3483 | 1.232 | 0.4751 | Yes | ||
| 45 | NDUFB7 | 3552 | 1.208 | 0.4806 | Yes | ||
| 46 | ATP4A | 3731 | 1.161 | 0.4798 | No | ||
| 47 | ATP5H | 4223 | 1.043 | 0.4612 | No | ||
| 48 | ATP5A1 | 4302 | 1.027 | 0.4648 | No | ||
| 49 | ATP5O | 4500 | 0.978 | 0.4616 | No | ||
| 50 | COX7B | 4536 | 0.970 | 0.4671 | No | ||
| 51 | ATP6V1E1 | 4806 | 0.913 | 0.4595 | No | ||
| 52 | COX7C | 4857 | 0.904 | 0.4637 | No | ||
| 53 | NDUFC1 | 5385 | 0.791 | 0.4412 | No | ||
| 54 | COX4I1 | 5551 | 0.759 | 0.4381 | No | ||
| 55 | NDUFA8 | 5674 | 0.730 | 0.4371 | No | ||
| 56 | ATP6V0A2 | 5718 | 0.721 | 0.4402 | No | ||
| 57 | ATP6V0C | 5818 | 0.700 | 0.4402 | No | ||
| 58 | COX17 | 6114 | 0.644 | 0.4292 | No | ||
| 59 | SDHA | 6608 | 0.555 | 0.4068 | No | ||
| 60 | ATP5J2 | 6691 | 0.539 | 0.4064 | No | ||
| 61 | ATP6V1C1 | 7031 | 0.470 | 0.3917 | No | ||
| 62 | COX15 | 7262 | 0.422 | 0.3825 | No | ||
| 63 | ATP6V1B1 | 7573 | 0.372 | 0.3686 | No | ||
| 64 | COX8C | 7666 | 0.353 | 0.3663 | No | ||
| 65 | COX8A | 7883 | 0.314 | 0.3570 | No | ||
| 66 | ATP5G3 | 7901 | 0.310 | 0.3584 | No | ||
| 67 | ATP5C1 | 8581 | 0.193 | 0.3232 | No | ||
| 68 | ATP5G2 | 9205 | 0.083 | 0.2902 | No | ||
| 69 | ATP6V1E2 | 9748 | -0.020 | 0.2611 | No | ||
| 70 | NDUFB11 | 9833 | -0.038 | 0.2568 | No | ||
| 71 | UQCR | 10121 | -0.089 | 0.2420 | No | ||
| 72 | ATP6V1G1 | 10124 | -0.089 | 0.2426 | No | ||
| 73 | ATP6V0D2 | 10275 | -0.116 | 0.2354 | No | ||
| 74 | NDUFS7 | 10279 | -0.116 | 0.2361 | No | ||
| 75 | COX4I2 | 10471 | -0.152 | 0.2269 | No | ||
| 76 | ATP4B | 10488 | -0.155 | 0.2272 | No | ||
| 77 | UQCRC1 | 10851 | -0.220 | 0.2094 | No | ||
| 78 | UQCRC2 | 10913 | -0.231 | 0.2078 | No | ||
| 79 | ATP6V0B | 11052 | -0.255 | 0.2023 | No | ||
| 80 | NDUFS1 | 11519 | -0.347 | 0.1798 | No | ||
| 81 | NDUFAB1 | 11655 | -0.378 | 0.1754 | No | ||
| 82 | ATP6V1G3 | 11701 | -0.387 | 0.1759 | No | ||
| 83 | ATP6V0A1 | 11977 | -0.435 | 0.1643 | No | ||
| 84 | COX6B2 | 12487 | -0.537 | 0.1409 | No | ||
| 85 | ATP5G1 | 12942 | -0.627 | 0.1212 | No | ||
| 86 | COX10 | 13699 | -0.795 | 0.0864 | No | ||
| 87 | ATP6V1C2 | 13779 | -0.815 | 0.0883 | No | ||
| 88 | NDUFS5 | 14394 | -0.972 | 0.0625 | No | ||
| 89 | NDUFV1 | 14449 | -0.986 | 0.0671 | No | ||
| 90 | COX5A | 14810 | -1.085 | 0.0559 | No | ||
| 91 | CYC1 | 15038 | -1.145 | 0.0524 | No | ||
| 92 | COX6A1 | 15131 | -1.174 | 0.0563 | No | ||
| 93 | ATP6V0A4 | 15147 | -1.178 | 0.0645 | No | ||
| 94 | ATP5D | 15308 | -1.222 | 0.0651 | No | ||
| 95 | COX6A2 | 15357 | -1.239 | 0.0720 | No | ||
| 96 | COX5B | 15464 | -1.274 | 0.0759 | No | ||
| 97 | ATP5B | 16165 | -1.538 | 0.0498 | No | ||
| 98 | ATP5J | 16352 | -1.605 | 0.0520 | No | ||
| 99 | TCIRG1 | 17945 | -2.899 | -0.0120 | No | ||
| 100 | ATP6V1G2 | 18040 | -3.030 | 0.0060 | No | ||
| 101 | NDUFA10 | 18197 | -3.299 | 0.0226 | No |