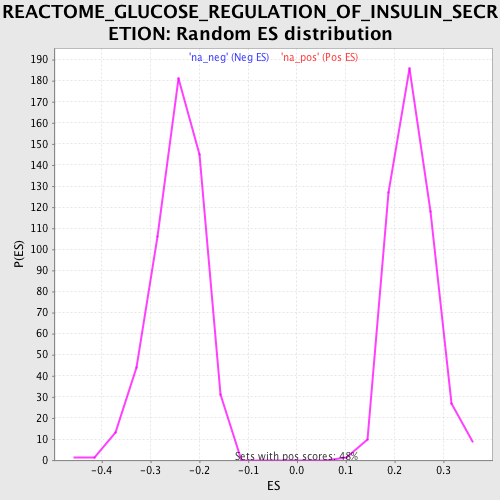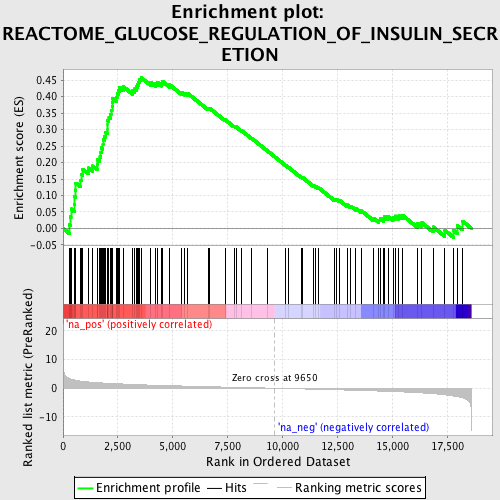
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_GLUCOSE_REGULATION_OF_INSULIN_SECRETION |
| Enrichment Score (ES) | 0.45756504 |
| Normalized Enrichment Score (NES) | 1.9508166 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.041309655 |
| FWER p-Value | 0.241 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COX7A2L | 290 | 3.370 | 0.0113 | Yes | ||
| 2 | CS | 324 | 3.290 | 0.0358 | Yes | ||
| 3 | NDUFA6 | 361 | 3.163 | 0.0592 | Yes | ||
| 4 | NDUFB3 | 512 | 2.877 | 0.0741 | Yes | ||
| 5 | SLC2A2 | 523 | 2.861 | 0.0965 | Yes | ||
| 6 | UQCRH | 562 | 2.785 | 0.1167 | Yes | ||
| 7 | SDHB | 581 | 2.756 | 0.1378 | Yes | ||
| 8 | NDUFA1 | 801 | 2.490 | 0.1459 | Yes | ||
| 9 | NDUFS4 | 831 | 2.449 | 0.1639 | Yes | ||
| 10 | NDUFB5 | 888 | 2.387 | 0.1800 | Yes | ||
| 11 | ATP5F1 | 1148 | 2.190 | 0.1835 | Yes | ||
| 12 | NDUFB4 | 1344 | 2.072 | 0.1896 | Yes | ||
| 13 | NDUFS3 | 1549 | 1.946 | 0.1941 | Yes | ||
| 14 | UQCRFS1 | 1579 | 1.930 | 0.2080 | Yes | ||
| 15 | CYCS | 1680 | 1.874 | 0.2176 | Yes | ||
| 16 | UQCRB | 1719 | 1.859 | 0.2304 | Yes | ||
| 17 | NDUFB8 | 1735 | 1.850 | 0.2444 | Yes | ||
| 18 | NDUFA11 | 1817 | 1.815 | 0.2546 | Yes | ||
| 19 | PGK1 | 1822 | 1.812 | 0.2689 | Yes | ||
| 20 | NDUFA5 | 1887 | 1.779 | 0.2796 | Yes | ||
| 21 | ATP5E | 1934 | 1.754 | 0.2912 | Yes | ||
| 22 | ETFB | 2013 | 1.718 | 0.3007 | Yes | ||
| 23 | NDUFC2 | 2016 | 1.717 | 0.3143 | Yes | ||
| 24 | NDUFB2 | 2018 | 1.716 | 0.3280 | Yes | ||
| 25 | PFKFB3 | 2089 | 1.689 | 0.3378 | Yes | ||
| 26 | NDUFA2 | 2164 | 1.660 | 0.3470 | Yes | ||
| 27 | COX6C | 2186 | 1.653 | 0.3591 | Yes | ||
| 28 | SDHD | 2230 | 1.637 | 0.3699 | Yes | ||
| 29 | COX6B1 | 2239 | 1.635 | 0.3826 | Yes | ||
| 30 | NDUFA7 | 2256 | 1.629 | 0.3947 | Yes | ||
| 31 | SYT5 | 2425 | 1.570 | 0.3982 | Yes | ||
| 32 | DLAT | 2462 | 1.557 | 0.4087 | Yes | ||
| 33 | NDUFA9 | 2510 | 1.543 | 0.4185 | Yes | ||
| 34 | NDUFA4 | 2561 | 1.527 | 0.4281 | Yes | ||
| 35 | ATP5I | 2735 | 1.471 | 0.4305 | Yes | ||
| 36 | NDUFS8 | 3177 | 1.326 | 0.4173 | Yes | ||
| 37 | NDUFA3 | 3263 | 1.301 | 0.4231 | Yes | ||
| 38 | NDUFS2 | 3336 | 1.281 | 0.4295 | Yes | ||
| 39 | OGDH | 3395 | 1.258 | 0.4364 | Yes | ||
| 40 | MDH2 | 3443 | 1.244 | 0.4438 | Yes | ||
| 41 | NDUFB10 | 3483 | 1.232 | 0.4516 | Yes | ||
| 42 | NDUFB7 | 3552 | 1.208 | 0.4576 | Yes | ||
| 43 | DLD | 3985 | 1.097 | 0.4430 | No | ||
| 44 | ATP5H | 4223 | 1.043 | 0.4386 | No | ||
| 45 | ATP5A1 | 4302 | 1.027 | 0.4426 | No | ||
| 46 | ATP5O | 4500 | 0.978 | 0.4398 | No | ||
| 47 | COX7B | 4536 | 0.970 | 0.4456 | No | ||
| 48 | COX7C | 4857 | 0.904 | 0.4356 | No | ||
| 49 | NDUFC1 | 5385 | 0.791 | 0.4135 | No | ||
| 50 | COX4I1 | 5551 | 0.759 | 0.4106 | No | ||
| 51 | NDUFA8 | 5674 | 0.730 | 0.4099 | No | ||
| 52 | SDHA | 6608 | 0.555 | 0.3640 | No | ||
| 53 | ATP5J2 | 6691 | 0.539 | 0.3639 | No | ||
| 54 | PFKFB1 | 7378 | 0.405 | 0.3301 | No | ||
| 55 | PDHX | 7791 | 0.332 | 0.3105 | No | ||
| 56 | COX8A | 7883 | 0.314 | 0.3081 | No | ||
| 57 | DLST | 8133 | 0.270 | 0.2968 | No | ||
| 58 | ATP5C1 | 8581 | 0.193 | 0.2742 | No | ||
| 59 | PRKACB | 9310 | 0.065 | 0.2355 | No | ||
| 60 | UQCR | 10121 | -0.089 | 0.1925 | No | ||
| 61 | NDUFS7 | 10279 | -0.116 | 0.1849 | No | ||
| 62 | UQCRC1 | 10851 | -0.220 | 0.1558 | No | ||
| 63 | UQCRC2 | 10913 | -0.231 | 0.1544 | No | ||
| 64 | IDH3A | 11413 | -0.327 | 0.1301 | No | ||
| 65 | NDUFS1 | 11519 | -0.347 | 0.1272 | No | ||
| 66 | NDUFAB1 | 11655 | -0.378 | 0.1229 | No | ||
| 67 | KCNJ11 | 12381 | -0.514 | 0.0879 | No | ||
| 68 | SUCLG1 | 12438 | -0.527 | 0.0891 | No | ||
| 69 | PDHA1 | 12598 | -0.559 | 0.0850 | No | ||
| 70 | ATP5G1 | 12942 | -0.627 | 0.0715 | No | ||
| 71 | ABCC8 | 13107 | -0.661 | 0.0679 | No | ||
| 72 | IDH3G | 13341 | -0.712 | 0.0611 | No | ||
| 73 | PFKFB2 | 13601 | -0.770 | 0.0532 | No | ||
| 74 | PDHB | 14167 | -0.911 | 0.0300 | No | ||
| 75 | NDUFS5 | 14394 | -0.972 | 0.0256 | No | ||
| 76 | NDUFV1 | 14449 | -0.986 | 0.0306 | No | ||
| 77 | ETFDH | 14613 | -1.030 | 0.0300 | No | ||
| 78 | IDH3B | 14659 | -1.042 | 0.0359 | No | ||
| 79 | COX5A | 14810 | -1.085 | 0.0365 | No | ||
| 80 | CYC1 | 15038 | -1.145 | 0.0334 | No | ||
| 81 | COX6A1 | 15131 | -1.174 | 0.0378 | No | ||
| 82 | ATP5D | 15308 | -1.222 | 0.0381 | No | ||
| 83 | COX5B | 15464 | -1.274 | 0.0400 | No | ||
| 84 | ATP5B | 16165 | -1.538 | 0.0145 | No | ||
| 85 | ATP5J | 16352 | -1.605 | 0.0173 | No | ||
| 86 | PKM2 | 16865 | -1.869 | 0.0046 | No | ||
| 87 | TPI1 | 17400 | -2.286 | -0.0059 | No | ||
| 88 | ETFA | 17796 | -2.702 | -0.0056 | No | ||
| 89 | VAMP2 | 17961 | -2.926 | 0.0089 | No | ||
| 90 | NDUFA10 | 18197 | -3.299 | 0.0226 | No |