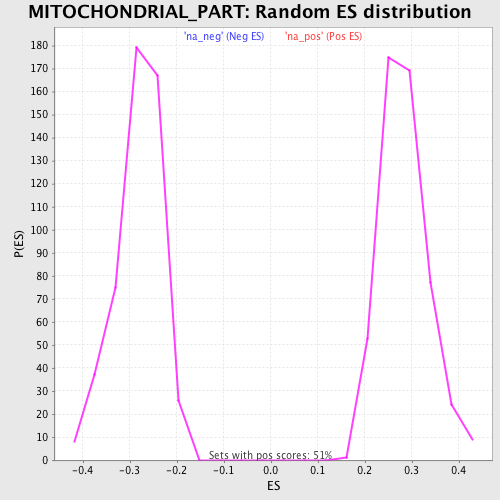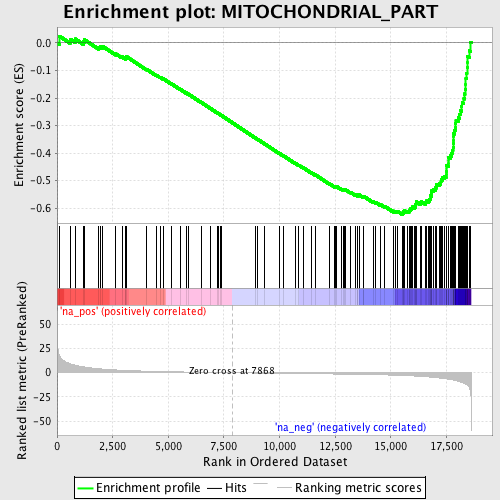
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | -0.6230791 |
| Normalized Enrichment Score (NES) | -2.2155042 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAB11FIP5 | 99 | 18.018 | 0.0249 | No | ||
| 2 | CASP7 | 584 | 9.126 | 0.0140 | No | ||
| 3 | COX6B2 | 811 | 7.682 | 0.0147 | No | ||
| 4 | SLC25A11 | 1189 | 5.957 | 0.0043 | No | ||
| 5 | MCL1 | 1227 | 5.811 | 0.0121 | No | ||
| 6 | ETFB | 1870 | 4.095 | -0.0158 | No | ||
| 7 | TIMM17B | 1955 | 3.913 | -0.0137 | No | ||
| 8 | SLC25A1 | 2050 | 3.728 | -0.0126 | No | ||
| 9 | MFN2 | 2630 | 2.818 | -0.0392 | No | ||
| 10 | CENTA2 | 2916 | 2.485 | -0.0504 | No | ||
| 11 | NR3C1 | 3088 | 2.317 | -0.0558 | No | ||
| 12 | HSD3B2 | 3090 | 2.316 | -0.0519 | No | ||
| 13 | NDUFA13 | 3115 | 2.288 | -0.0494 | No | ||
| 14 | HCCS | 4020 | 1.568 | -0.0956 | No | ||
| 15 | SDHA | 4451 | 1.310 | -0.1167 | No | ||
| 16 | UQCRH | 4640 | 1.202 | -0.1248 | No | ||
| 17 | ALAS2 | 4765 | 1.148 | -0.1296 | No | ||
| 18 | UQCRB | 5134 | 0.977 | -0.1479 | No | ||
| 19 | SLC25A22 | 5544 | 0.795 | -0.1687 | No | ||
| 20 | RAF1 | 5794 | 0.702 | -0.1810 | No | ||
| 21 | NDUFS4 | 5894 | 0.667 | -0.1852 | No | ||
| 22 | NDUFA6 | 6467 | 0.445 | -0.2154 | No | ||
| 23 | VDAC1 | 6909 | 0.294 | -0.2388 | No | ||
| 24 | MTX2 | 7214 | 0.197 | -0.2549 | No | ||
| 25 | PHB | 7255 | 0.180 | -0.2567 | No | ||
| 26 | MRPS22 | 7329 | 0.158 | -0.2604 | No | ||
| 27 | GATM | 7410 | 0.139 | -0.2645 | No | ||
| 28 | NDUFS1 | 8907 | -0.316 | -0.3449 | No | ||
| 29 | NDUFA1 | 9024 | -0.348 | -0.3506 | No | ||
| 30 | PITRM1 | 9333 | -0.438 | -0.3665 | No | ||
| 31 | MAOB | 10013 | -0.633 | -0.4022 | No | ||
| 32 | MRPS36 | 10158 | -0.671 | -0.4088 | No | ||
| 33 | PPOX | 10706 | -0.826 | -0.4370 | No | ||
| 34 | RHOT2 | 10866 | -0.868 | -0.4442 | No | ||
| 35 | SLC25A3 | 11062 | -0.919 | -0.4532 | No | ||
| 36 | MRPL32 | 11444 | -1.031 | -0.4721 | No | ||
| 37 | NDUFS8 | 11611 | -1.076 | -0.4792 | No | ||
| 38 | ATP5C1 | 12257 | -1.274 | -0.5120 | No | ||
| 39 | OGDH | 12484 | -1.344 | -0.5219 | No | ||
| 40 | CASQ1 | 12512 | -1.352 | -0.5211 | No | ||
| 41 | ATP5B | 12557 | -1.363 | -0.5212 | No | ||
| 42 | PIN4 | 12782 | -1.439 | -0.5309 | No | ||
| 43 | BNIP3 | 12876 | -1.470 | -0.5335 | No | ||
| 44 | TIMM23 | 12922 | -1.485 | -0.5334 | No | ||
| 45 | NDUFA2 | 12942 | -1.490 | -0.5320 | No | ||
| 46 | UCP3 | 13187 | -1.571 | -0.5425 | No | ||
| 47 | MRPS16 | 13405 | -1.653 | -0.5515 | No | ||
| 48 | HSD3B1 | 13507 | -1.694 | -0.5541 | No | ||
| 49 | ATP5G2 | 13518 | -1.699 | -0.5518 | No | ||
| 50 | UQCRC1 | 13571 | -1.722 | -0.5517 | No | ||
| 51 | MRPL51 | 13767 | -1.797 | -0.5592 | No | ||
| 52 | FIS1 | 13771 | -1.799 | -0.5564 | No | ||
| 53 | RHOT1 | 14206 | -1.993 | -0.5765 | No | ||
| 54 | DBT | 14310 | -2.048 | -0.5786 | No | ||
| 55 | CS | 14525 | -2.160 | -0.5866 | No | ||
| 56 | MRPS18C | 14737 | -2.270 | -0.5942 | No | ||
| 57 | ATP5J | 15114 | -2.506 | -0.6103 | No | ||
| 58 | ATP5E | 15220 | -2.582 | -0.6117 | No | ||
| 59 | TIMM9 | 15291 | -2.637 | -0.6110 | No | ||
| 60 | TOMM34 | 15515 | -2.802 | -0.6184 | Yes | ||
| 61 | ABCB7 | 15568 | -2.850 | -0.6164 | Yes | ||
| 62 | ATP5A1 | 15576 | -2.855 | -0.6120 | Yes | ||
| 63 | MRPS21 | 15611 | -2.886 | -0.6090 | Yes | ||
| 64 | BCL2 | 15743 | -3.007 | -0.6110 | Yes | ||
| 65 | MPV17 | 15818 | -3.108 | -0.6098 | Yes | ||
| 66 | GRPEL1 | 15858 | -3.147 | -0.6066 | Yes | ||
| 67 | BCKDK | 15885 | -3.171 | -0.6027 | Yes | ||
| 68 | VDAC3 | 15936 | -3.232 | -0.6000 | Yes | ||
| 69 | NNT | 15954 | -3.247 | -0.5955 | Yes | ||
| 70 | ATP5O | 16055 | -3.362 | -0.5952 | Yes | ||
| 71 | ATP5G3 | 16106 | -3.421 | -0.5922 | Yes | ||
| 72 | BCKDHA | 16119 | -3.433 | -0.5871 | Yes | ||
| 73 | ATP5D | 16132 | -3.445 | -0.5819 | Yes | ||
| 74 | MRPL52 | 16151 | -3.474 | -0.5771 | Yes | ||
| 75 | ATP5F1 | 16323 | -3.717 | -0.5801 | Yes | ||
| 76 | OPA1 | 16360 | -3.773 | -0.5757 | Yes | ||
| 77 | TFB2M | 16564 | -4.101 | -0.5798 | Yes | ||
| 78 | NDUFS7 | 16583 | -4.134 | -0.5738 | Yes | ||
| 79 | COX15 | 16694 | -4.351 | -0.5725 | Yes | ||
| 80 | DNAJA3 | 16733 | -4.422 | -0.5671 | Yes | ||
| 81 | BCKDHB | 16780 | -4.532 | -0.5620 | Yes | ||
| 82 | ALDH4A1 | 16782 | -4.539 | -0.5544 | Yes | ||
| 83 | AIFM2 | 16825 | -4.628 | -0.5489 | Yes | ||
| 84 | SDHD | 16828 | -4.632 | -0.5412 | Yes | ||
| 85 | NDUFAB1 | 16844 | -4.660 | -0.5342 | Yes | ||
| 86 | MRPS15 | 16929 | -4.811 | -0.5307 | Yes | ||
| 87 | TIMM44 | 17005 | -4.960 | -0.5264 | Yes | ||
| 88 | SURF1 | 17034 | -5.031 | -0.5195 | Yes | ||
| 89 | ACADM | 17063 | -5.088 | -0.5125 | Yes | ||
| 90 | MRPL23 | 17176 | -5.414 | -0.5094 | Yes | ||
| 91 | HTRA2 | 17254 | -5.607 | -0.5042 | Yes | ||
| 92 | NDUFS2 | 17262 | -5.625 | -0.4951 | Yes | ||
| 93 | MRPL55 | 17319 | -5.752 | -0.4885 | Yes | ||
| 94 | TOMM22 | 17419 | -6.052 | -0.4837 | Yes | ||
| 95 | MRPS28 | 17486 | -6.297 | -0.4767 | Yes | ||
| 96 | TIMM8B | 17491 | -6.344 | -0.4662 | Yes | ||
| 97 | IMMT | 17501 | -6.374 | -0.4560 | Yes | ||
| 98 | ETFA | 17502 | -6.381 | -0.4453 | Yes | ||
| 99 | ACN9 | 17591 | -6.678 | -0.4389 | Yes | ||
| 100 | NDUFV1 | 17595 | -6.695 | -0.4278 | Yes | ||
| 101 | MRPS12 | 17597 | -6.703 | -0.4166 | Yes | ||
| 102 | PMPCA | 17677 | -7.018 | -0.4091 | Yes | ||
| 103 | HADHB | 17737 | -7.236 | -0.4001 | Yes | ||
| 104 | ATP5G1 | 17758 | -7.336 | -0.3889 | Yes | ||
| 105 | NDUFA9 | 17795 | -7.504 | -0.3782 | Yes | ||
| 106 | POLG2 | 17807 | -7.563 | -0.3661 | Yes | ||
| 107 | MRPS18A | 17816 | -7.611 | -0.3538 | Yes | ||
| 108 | MRPS11 | 17822 | -7.639 | -0.3412 | Yes | ||
| 109 | TIMM50 | 17828 | -7.647 | -0.3287 | Yes | ||
| 110 | ABCB6 | 17872 | -7.856 | -0.3178 | Yes | ||
| 111 | VDAC2 | 17896 | -7.962 | -0.3057 | Yes | ||
| 112 | MRPL40 | 17906 | -7.977 | -0.2928 | Yes | ||
| 113 | TIMM10 | 17928 | -8.102 | -0.2803 | Yes | ||
| 114 | MRPL10 | 18029 | -8.693 | -0.2711 | Yes | ||
| 115 | CYCS | 18087 | -9.108 | -0.2589 | Yes | ||
| 116 | BCS1L | 18120 | -9.344 | -0.2449 | Yes | ||
| 117 | SUPV3L1 | 18172 | -9.846 | -0.2312 | Yes | ||
| 118 | MRPS24 | 18199 | -10.028 | -0.2157 | Yes | ||
| 119 | MRPS35 | 18286 | -10.769 | -0.2023 | Yes | ||
| 120 | NDUFS3 | 18313 | -11.057 | -0.1851 | Yes | ||
| 121 | TIMM17A | 18335 | -11.336 | -0.1672 | Yes | ||
| 122 | NFS1 | 18370 | -11.854 | -0.1492 | Yes | ||
| 123 | ABCF2 | 18376 | -11.997 | -0.1293 | Yes | ||
| 124 | PHB2 | 18395 | -12.155 | -0.1099 | Yes | ||
| 125 | SLC25A15 | 18425 | -12.538 | -0.0904 | Yes | ||
| 126 | BAX | 18434 | -12.657 | -0.0696 | Yes | ||
| 127 | MRPL12 | 18440 | -12.794 | -0.0484 | Yes | ||
| 128 | MRPS10 | 18528 | -15.055 | -0.0278 | Yes | ||
| 129 | TIMM13 | 18582 | -19.357 | 0.0018 | Yes |