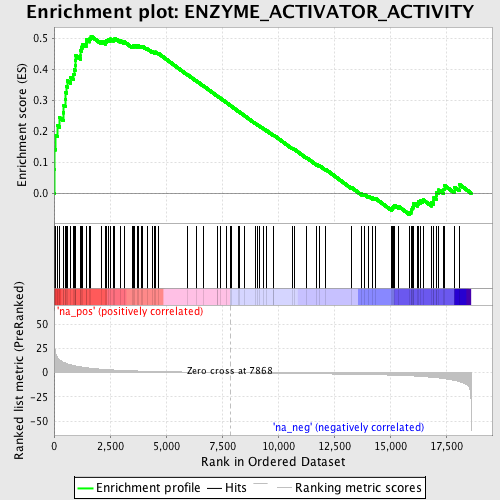
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ENZYME_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | 0.50746274 |
| Normalized Enrichment Score (NES) | 1.7340504 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17442842 |
| FWER p-Value | 0.942 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0770 | Yes | ||
| 2 | ABR | 21 | 28.176 | 0.1418 | Yes | ||
| 3 | PRKRA | 66 | 20.547 | 0.1870 | Yes | ||
| 4 | GMFG | 148 | 15.439 | 0.2184 | Yes | ||
| 5 | THY1 | 245 | 13.250 | 0.2439 | Yes | ||
| 6 | ARFGAP3 | 410 | 10.819 | 0.2601 | Yes | ||
| 7 | RASA3 | 433 | 10.636 | 0.2836 | Yes | ||
| 8 | SIPA1 | 501 | 9.928 | 0.3029 | Yes | ||
| 9 | BCR | 517 | 9.784 | 0.3248 | Yes | ||
| 10 | ALOX5AP | 559 | 9.304 | 0.3441 | Yes | ||
| 11 | RALBP1 | 597 | 9.033 | 0.3631 | Yes | ||
| 12 | NOD1 | 741 | 8.108 | 0.3741 | Yes | ||
| 13 | RASA4 | 868 | 7.374 | 0.3844 | Yes | ||
| 14 | PSAP | 909 | 7.163 | 0.3988 | Yes | ||
| 15 | MAP3K5 | 939 | 7.023 | 0.4135 | Yes | ||
| 16 | RGS12 | 950 | 6.962 | 0.4291 | Yes | ||
| 17 | ARHGAP4 | 967 | 6.888 | 0.4442 | Yes | ||
| 18 | GUCA1A | 1183 | 5.978 | 0.4465 | Yes | ||
| 19 | CDC42EP2 | 1188 | 5.960 | 0.4600 | Yes | ||
| 20 | MYO9B | 1213 | 5.867 | 0.4723 | Yes | ||
| 21 | PRKCD | 1279 | 5.615 | 0.4818 | Yes | ||
| 22 | RASA1 | 1431 | 5.144 | 0.4856 | Yes | ||
| 23 | FZR1 | 1450 | 5.087 | 0.4964 | Yes | ||
| 24 | CASP1 | 1599 | 4.688 | 0.4993 | Yes | ||
| 25 | ARHGAP10 | 1645 | 4.583 | 0.5075 | Yes | ||
| 26 | RGS1 | 2112 | 3.615 | 0.4907 | No | ||
| 27 | OPHN1 | 2283 | 3.333 | 0.4892 | No | ||
| 28 | RGS2 | 2330 | 3.259 | 0.4943 | No | ||
| 29 | GUCA1C | 2430 | 3.104 | 0.4961 | No | ||
| 30 | RABGAP1 | 2518 | 2.986 | 0.4983 | No | ||
| 31 | CENTD2 | 2648 | 2.799 | 0.4979 | No | ||
| 32 | GHRL | 2713 | 2.717 | 0.5007 | No | ||
| 33 | CHN2 | 2957 | 2.437 | 0.4932 | No | ||
| 34 | NF1 | 3132 | 2.270 | 0.4891 | No | ||
| 35 | MADD | 3479 | 1.951 | 0.4749 | No | ||
| 36 | IGFBP3 | 3539 | 1.907 | 0.4761 | No | ||
| 37 | RCVRN | 3587 | 1.874 | 0.4779 | No | ||
| 38 | RGS9 | 3741 | 1.754 | 0.4737 | No | ||
| 39 | DNMT3L | 3755 | 1.742 | 0.4771 | No | ||
| 40 | ALS2 | 3900 | 1.637 | 0.4731 | No | ||
| 41 | HSPB2 | 3946 | 1.612 | 0.4744 | No | ||
| 42 | F10 | 4163 | 1.495 | 0.4662 | No | ||
| 43 | RGS6 | 4393 | 1.353 | 0.4570 | No | ||
| 44 | FGF13 | 4501 | 1.284 | 0.4541 | No | ||
| 45 | RASA2 | 4517 | 1.270 | 0.4563 | No | ||
| 46 | SOS1 | 4644 | 1.201 | 0.4523 | No | ||
| 47 | TSC2 | 5954 | 0.645 | 0.3831 | No | ||
| 48 | MMP15 | 6346 | 0.488 | 0.3631 | No | ||
| 49 | RGS11 | 6673 | 0.375 | 0.3463 | No | ||
| 50 | ARFGEF1 | 7308 | 0.166 | 0.3125 | No | ||
| 51 | BCL10 | 7310 | 0.166 | 0.3128 | No | ||
| 52 | GUCA1B | 7428 | 0.133 | 0.3068 | No | ||
| 53 | CASP8AP2 | 7699 | 0.048 | 0.2923 | No | ||
| 54 | APOA5 | 7860 | 0.003 | 0.2837 | No | ||
| 55 | APOA4 | 7938 | -0.019 | 0.2796 | No | ||
| 56 | DEPDC2 | 8242 | -0.116 | 0.2635 | No | ||
| 57 | RFC1 | 8267 | -0.124 | 0.2625 | No | ||
| 58 | RASGRP3 | 8508 | -0.196 | 0.2500 | No | ||
| 59 | ALDH1A1 | 8973 | -0.335 | 0.2257 | No | ||
| 60 | GMIP | 9085 | -0.366 | 0.2205 | No | ||
| 61 | RGS14 | 9172 | -0.391 | 0.2168 | No | ||
| 62 | PITRM1 | 9333 | -0.438 | 0.2092 | No | ||
| 63 | MMP16 | 9500 | -0.486 | 0.2013 | No | ||
| 64 | TPD52L1 | 9799 | -0.572 | 0.1865 | No | ||
| 65 | CXCL1 | 9802 | -0.573 | 0.1878 | No | ||
| 66 | APOC2 | 10633 | -0.804 | 0.1448 | No | ||
| 67 | DNAJC1 | 10730 | -0.834 | 0.1416 | No | ||
| 68 | MMP24 | 11259 | -0.974 | 0.1153 | No | ||
| 69 | IL2 | 11727 | -1.116 | 0.0927 | No | ||
| 70 | MAPK8IP2 | 11852 | -1.156 | 0.0886 | No | ||
| 71 | RACGAP1 | 12113 | -1.234 | 0.0774 | No | ||
| 72 | ARL1 | 13256 | -1.595 | 0.0195 | No | ||
| 73 | APOA2 | 13731 | -1.783 | -0.0020 | No | ||
| 74 | ARHGAP5 | 13857 | -1.836 | -0.0045 | No | ||
| 75 | GUCA2B | 14034 | -1.914 | -0.0096 | No | ||
| 76 | PPP2R4 | 14217 | -1.995 | -0.0148 | No | ||
| 77 | PLAA | 14324 | -2.053 | -0.0158 | No | ||
| 78 | DLC1 | 15061 | -2.463 | -0.0498 | No | ||
| 79 | DOCK4 | 15119 | -2.509 | -0.0471 | No | ||
| 80 | RGS20 | 15136 | -2.521 | -0.0421 | No | ||
| 81 | MMP17 | 15196 | -2.566 | -0.0393 | No | ||
| 82 | RGS4 | 15388 | -2.709 | -0.0434 | No | ||
| 83 | CASP9 | 15860 | -3.149 | -0.0615 | No | ||
| 84 | BNIP2 | 15933 | -3.231 | -0.0579 | No | ||
| 85 | GMFB | 15969 | -3.264 | -0.0522 | No | ||
| 86 | RASAL1 | 15979 | -3.275 | -0.0451 | No | ||
| 87 | RANGAP1 | 16032 | -3.337 | -0.0402 | No | ||
| 88 | MALT1 | 16052 | -3.358 | -0.0335 | No | ||
| 89 | DNAJB6 | 16207 | -3.555 | -0.0336 | No | ||
| 90 | RGS3 | 16253 | -3.614 | -0.0276 | No | ||
| 91 | DBNL | 16363 | -3.776 | -0.0248 | No | ||
| 92 | TRIM23 | 16467 | -3.950 | -0.0212 | No | ||
| 93 | CENTA1 | 16847 | -4.663 | -0.0308 | No | ||
| 94 | ADRM1 | 16948 | -4.841 | -0.0250 | No | ||
| 95 | AHSA1 | 16953 | -4.851 | -0.0140 | No | ||
| 96 | CD24 | 17062 | -5.087 | -0.0080 | No | ||
| 97 | RGS16 | 17080 | -5.145 | 0.0030 | No | ||
| 98 | APOA1 | 17173 | -5.406 | 0.0105 | No | ||
| 99 | CTAGE5 | 17397 | -6.002 | 0.0124 | No | ||
| 100 | CTSA | 17434 | -6.097 | 0.0246 | No | ||
| 101 | GTF3C4 | 17858 | -7.795 | 0.0198 | No | ||
| 102 | CDK5R1 | 18088 | -9.118 | 0.0285 | No |
