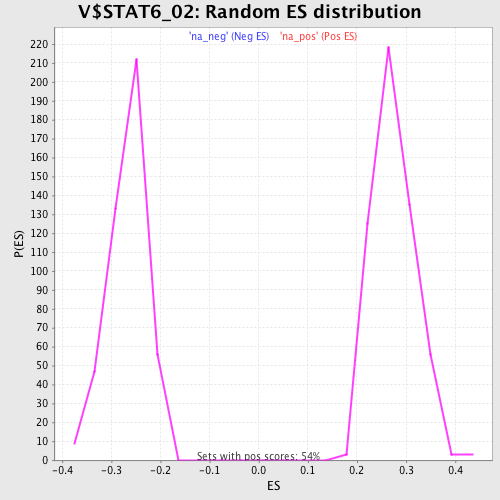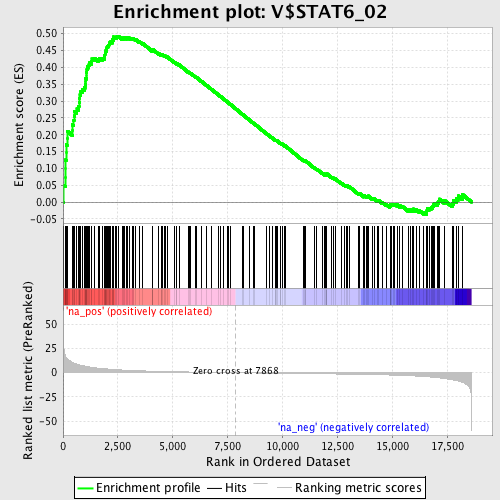
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$STAT6_02 |
| Enrichment Score (ES) | 0.49283192 |
| Normalized Enrichment Score (NES) | 1.7945727 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05870125 |
| FWER p-Value | 0.172 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0497 | Yes | ||
| 2 | EGFL7 | 100 | 17.985 | 0.0719 | Yes | ||
| 3 | SYTL1 | 103 | 17.817 | 0.0986 | Yes | ||
| 4 | CA3 | 105 | 17.758 | 0.1252 | Yes | ||
| 5 | SATB1 | 140 | 15.729 | 0.1470 | Yes | ||
| 6 | SGK | 143 | 15.627 | 0.1703 | Yes | ||
| 7 | MEF2C | 185 | 14.237 | 0.1895 | Yes | ||
| 8 | HHEX | 190 | 14.145 | 0.2105 | Yes | ||
| 9 | ERG | 426 | 10.702 | 0.2138 | Yes | ||
| 10 | LAT | 432 | 10.644 | 0.2295 | Yes | ||
| 11 | MAP3K3 | 488 | 10.054 | 0.2416 | Yes | ||
| 12 | SIPA1 | 501 | 9.928 | 0.2559 | Yes | ||
| 13 | PDE4D | 524 | 9.718 | 0.2693 | Yes | ||
| 14 | IL7R | 631 | 8.739 | 0.2767 | Yes | ||
| 15 | MAP4K2 | 708 | 8.299 | 0.2850 | Yes | ||
| 16 | PDLIM2 | 729 | 8.184 | 0.2962 | Yes | ||
| 17 | CTCF | 758 | 8.038 | 0.3068 | Yes | ||
| 18 | HOXB4 | 768 | 7.921 | 0.3182 | Yes | ||
| 19 | STAT5A | 807 | 7.710 | 0.3277 | Yes | ||
| 20 | NFAT5 | 871 | 7.334 | 0.3353 | Yes | ||
| 21 | MBNL1 | 963 | 6.901 | 0.3407 | Yes | ||
| 22 | GNB2 | 1001 | 6.736 | 0.3488 | Yes | ||
| 23 | PRKCB1 | 1028 | 6.604 | 0.3573 | Yes | ||
| 24 | ABI3 | 1032 | 6.597 | 0.3671 | Yes | ||
| 25 | ZRANB1 | 1068 | 6.481 | 0.3749 | Yes | ||
| 26 | GPHN | 1070 | 6.470 | 0.3846 | Yes | ||
| 27 | CASKIN2 | 1085 | 6.396 | 0.3934 | Yes | ||
| 28 | HSD11B1 | 1105 | 6.325 | 0.4019 | Yes | ||
| 29 | LCK | 1176 | 6.008 | 0.4071 | Yes | ||
| 30 | ZDHHC8 | 1217 | 5.849 | 0.4137 | Yes | ||
| 31 | RIN2 | 1296 | 5.538 | 0.4178 | Yes | ||
| 32 | LTC4S | 1315 | 5.478 | 0.4250 | Yes | ||
| 33 | SLC38A2 | 1421 | 5.175 | 0.4271 | Yes | ||
| 34 | HNRPUL1 | 1600 | 4.688 | 0.4245 | Yes | ||
| 35 | CAMK1D | 1666 | 4.519 | 0.4278 | Yes | ||
| 36 | NOTCH4 | 1784 | 4.256 | 0.4278 | Yes | ||
| 37 | PHF12 | 1877 | 4.077 | 0.4289 | Yes | ||
| 38 | PITPNC1 | 1880 | 4.070 | 0.4350 | Yes | ||
| 39 | CDKL5 | 1909 | 4.007 | 0.4394 | Yes | ||
| 40 | EGF | 1914 | 3.994 | 0.4452 | Yes | ||
| 41 | FEM1C | 1950 | 3.923 | 0.4492 | Yes | ||
| 42 | DCTN2 | 1975 | 3.877 | 0.4537 | Yes | ||
| 43 | STK4 | 1996 | 3.829 | 0.4584 | Yes | ||
| 44 | USP2 | 2010 | 3.801 | 0.4634 | Yes | ||
| 45 | TCF15 | 2065 | 3.695 | 0.4660 | Yes | ||
| 46 | ITPR3 | 2092 | 3.648 | 0.4701 | Yes | ||
| 47 | PDGFRB | 2130 | 3.579 | 0.4735 | Yes | ||
| 48 | RANBP9 | 2176 | 3.510 | 0.4763 | Yes | ||
| 49 | ARID1A | 2251 | 3.402 | 0.4774 | Yes | ||
| 50 | MPO | 2256 | 3.396 | 0.4823 | Yes | ||
| 51 | LAG3 | 2290 | 3.316 | 0.4855 | Yes | ||
| 52 | CNN2 | 2291 | 3.314 | 0.4904 | Yes | ||
| 53 | ZNF289 | 2403 | 3.137 | 0.4891 | Yes | ||
| 54 | BZRAP1 | 2450 | 3.077 | 0.4913 | Yes | ||
| 55 | SDCBP | 2505 | 3.002 | 0.4928 | Yes | ||
| 56 | VAMP3 | 2691 | 2.745 | 0.4869 | No | ||
| 57 | CPA3 | 2751 | 2.683 | 0.4877 | No | ||
| 58 | TNFSF13 | 2800 | 2.629 | 0.4891 | No | ||
| 59 | GPS2 | 2898 | 2.502 | 0.4876 | No | ||
| 60 | LNX2 | 2942 | 2.455 | 0.4889 | No | ||
| 61 | CSF2 | 3037 | 2.356 | 0.4874 | No | ||
| 62 | CYB561D2 | 3150 | 2.246 | 0.4847 | No | ||
| 63 | CSF3R | 3189 | 2.206 | 0.4859 | No | ||
| 64 | PLCB2 | 3305 | 2.098 | 0.4828 | No | ||
| 65 | JARID1C | 3467 | 1.957 | 0.4771 | No | ||
| 66 | TRIM25 | 3626 | 1.837 | 0.4712 | No | ||
| 67 | STRC | 4067 | 1.544 | 0.4497 | No | ||
| 68 | PHF15 | 4070 | 1.543 | 0.4519 | No | ||
| 69 | SRRM1 | 4088 | 1.531 | 0.4533 | No | ||
| 70 | PSCD4 | 4354 | 1.375 | 0.4410 | No | ||
| 71 | HGFAC | 4477 | 1.300 | 0.4363 | No | ||
| 72 | FGF13 | 4501 | 1.284 | 0.4370 | No | ||
| 73 | STAB1 | 4515 | 1.272 | 0.4382 | No | ||
| 74 | RBBP5 | 4625 | 1.211 | 0.4341 | No | ||
| 75 | HIVEP1 | 4662 | 1.194 | 0.4340 | No | ||
| 76 | TUSC4 | 4746 | 1.158 | 0.4312 | No | ||
| 77 | DHH | 5073 | 1.004 | 0.4150 | No | ||
| 78 | TFEC | 5173 | 0.961 | 0.4111 | No | ||
| 79 | MRPL24 | 5178 | 0.957 | 0.4123 | No | ||
| 80 | VPS39 | 5285 | 0.905 | 0.4079 | No | ||
| 81 | FLI1 | 5702 | 0.729 | 0.3865 | No | ||
| 82 | WNT5A | 5763 | 0.709 | 0.3843 | No | ||
| 83 | LRP2 | 5817 | 0.694 | 0.3824 | No | ||
| 84 | PAX8 | 6047 | 0.604 | 0.3709 | No | ||
| 85 | EPHA7 | 6061 | 0.598 | 0.3711 | No | ||
| 86 | PGM2L1 | 6320 | 0.497 | 0.3579 | No | ||
| 87 | FBXW11 | 6527 | 0.426 | 0.3473 | No | ||
| 88 | SNX8 | 6757 | 0.347 | 0.3354 | No | ||
| 89 | PAX9 | 7080 | 0.239 | 0.3183 | No | ||
| 90 | RHOJ | 7159 | 0.216 | 0.3144 | No | ||
| 91 | GSC | 7302 | 0.169 | 0.3070 | No | ||
| 92 | BMP2 | 7510 | 0.111 | 0.2959 | No | ||
| 93 | EPHA2 | 7522 | 0.106 | 0.2955 | No | ||
| 94 | MLLT6 | 7613 | 0.078 | 0.2907 | No | ||
| 95 | CACNG3 | 8196 | -0.101 | 0.2593 | No | ||
| 96 | PDCD10 | 8204 | -0.102 | 0.2591 | No | ||
| 97 | GZMK | 8489 | -0.191 | 0.2440 | No | ||
| 98 | ITGA8 | 8669 | -0.246 | 0.2346 | No | ||
| 99 | TMC3 | 8721 | -0.262 | 0.2323 | No | ||
| 100 | CD2AP | 9269 | -0.421 | 0.2032 | No | ||
| 101 | MIST | 9406 | -0.459 | 0.1965 | No | ||
| 102 | HMGA1 | 9531 | -0.495 | 0.1906 | No | ||
| 103 | CRYGB | 9679 | -0.536 | 0.1834 | No | ||
| 104 | HOXB9 | 9688 | -0.538 | 0.1838 | No | ||
| 105 | RUNX3 | 9704 | -0.545 | 0.1838 | No | ||
| 106 | CITED2 | 9749 | -0.557 | 0.1822 | No | ||
| 107 | SCGB3A1 | 9907 | -0.604 | 0.1746 | No | ||
| 108 | GREM1 | 9928 | -0.609 | 0.1745 | No | ||
| 109 | LALBA | 9979 | -0.620 | 0.1727 | No | ||
| 110 | CMYA5 | 10104 | -0.660 | 0.1669 | No | ||
| 111 | SPDEF | 10115 | -0.661 | 0.1674 | No | ||
| 112 | TRAF3 | 10940 | -0.887 | 0.1240 | No | ||
| 113 | A2BP1 | 10979 | -0.897 | 0.1233 | No | ||
| 114 | TLL1 | 11029 | -0.912 | 0.1220 | No | ||
| 115 | PKN3 | 11051 | -0.917 | 0.1223 | No | ||
| 116 | PUM1 | 11469 | -1.036 | 0.1012 | No | ||
| 117 | HOXA2 | 11564 | -1.064 | 0.0977 | No | ||
| 118 | HTR3B | 11811 | -1.141 | 0.0861 | No | ||
| 119 | ERF | 11926 | -1.175 | 0.0817 | No | ||
| 120 | TOM1L1 | 11941 | -1.179 | 0.0827 | No | ||
| 121 | FEV | 12009 | -1.199 | 0.0808 | No | ||
| 122 | PSTPIP2 | 12010 | -1.200 | 0.0826 | No | ||
| 123 | ADCYAP1 | 12020 | -1.204 | 0.0840 | No | ||
| 124 | FLNC | 12245 | -1.271 | 0.0737 | No | ||
| 125 | GRIN2B | 12319 | -1.292 | 0.0717 | No | ||
| 126 | PPP2R5C | 12397 | -1.317 | 0.0695 | No | ||
| 127 | POU3F3 | 12677 | -1.403 | 0.0565 | No | ||
| 128 | LRRC19 | 12825 | -1.451 | 0.0507 | No | ||
| 129 | BDNF | 12898 | -1.477 | 0.0490 | No | ||
| 130 | TBC1D17 | 12939 | -1.489 | 0.0491 | No | ||
| 131 | ANGPTL1 | 13048 | -1.522 | 0.0455 | No | ||
| 132 | GPX1 | 13482 | -1.684 | 0.0246 | No | ||
| 133 | SLIT3 | 13510 | -1.695 | 0.0256 | No | ||
| 134 | GNGT2 | 13708 | -1.773 | 0.0176 | No | ||
| 135 | FLT1 | 13723 | -1.780 | 0.0195 | No | ||
| 136 | LY75 | 13839 | -1.828 | 0.0160 | No | ||
| 137 | RNF128 | 13895 | -1.852 | 0.0158 | No | ||
| 138 | SESTD1 | 13902 | -1.854 | 0.0183 | No | ||
| 139 | TANK | 14102 | -1.942 | 0.0104 | No | ||
| 140 | HIF3A | 14170 | -1.975 | 0.0097 | No | ||
| 141 | ASPN | 14321 | -2.052 | 0.0047 | No | ||
| 142 | TRPS1 | 14365 | -2.071 | 0.0055 | No | ||
| 143 | EDN1 | 14566 | -2.181 | -0.0021 | No | ||
| 144 | HOXA10 | 14722 | -2.262 | -0.0071 | No | ||
| 145 | AKT1S1 | 14898 | -2.361 | -0.0131 | No | ||
| 146 | NKIRAS2 | 14909 | -2.368 | -0.0100 | No | ||
| 147 | MCTS1 | 14930 | -2.386 | -0.0075 | No | ||
| 148 | SLC35C1 | 14978 | -2.412 | -0.0065 | No | ||
| 149 | DLC1 | 15061 | -2.463 | -0.0072 | No | ||
| 150 | DOCK4 | 15119 | -2.509 | -0.0065 | No | ||
| 151 | ATP5E | 15220 | -2.582 | -0.0081 | No | ||
| 152 | MTUS1 | 15353 | -2.685 | -0.0112 | No | ||
| 153 | ACBD6 | 15466 | -2.764 | -0.0131 | No | ||
| 154 | SLC31A1 | 15732 | -2.998 | -0.0230 | No | ||
| 155 | KCNQ1 | 15817 | -3.108 | -0.0229 | No | ||
| 156 | CLOCK | 15937 | -3.233 | -0.0245 | No | ||
| 157 | VASP | 15953 | -3.246 | -0.0204 | No | ||
| 158 | TRERF1 | 16095 | -3.410 | -0.0230 | No | ||
| 159 | RGS3 | 16253 | -3.614 | -0.0261 | No | ||
| 160 | CBLB | 16437 | -3.895 | -0.0301 | No | ||
| 161 | SEMA4B | 16562 | -4.098 | -0.0307 | No | ||
| 162 | SLC1A5 | 16582 | -4.130 | -0.0255 | No | ||
| 163 | SERPINI1 | 16587 | -4.143 | -0.0195 | No | ||
| 164 | TWIST1 | 16679 | -4.320 | -0.0180 | No | ||
| 165 | RNF138 | 16772 | -4.516 | -0.0162 | No | ||
| 166 | CENTA1 | 16847 | -4.663 | -0.0132 | No | ||
| 167 | CRK | 16873 | -4.709 | -0.0075 | No | ||
| 168 | SPI1 | 16932 | -4.818 | -0.0034 | No | ||
| 169 | DNAJC7 | 17057 | -5.082 | -0.0025 | No | ||
| 170 | POLR1D | 17100 | -5.209 | 0.0031 | No | ||
| 171 | LUC7L | 17136 | -5.315 | 0.0091 | No | ||
| 172 | TTC13 | 17369 | -5.923 | 0.0055 | No | ||
| 173 | DNAJA2 | 17734 | -7.231 | -0.0034 | No | ||
| 174 | COX17 | 17785 | -7.435 | 0.0050 | No | ||
| 175 | PIM2 | 17916 | -8.044 | 0.0101 | No | ||
| 176 | RAB39 | 18001 | -8.541 | 0.0183 | No | ||
| 177 | ADSL | 18193 | -9.970 | 0.0229 | No |