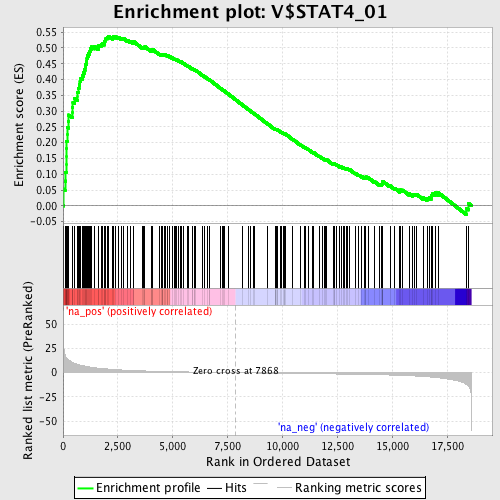
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$STAT4_01 |
| Enrichment Score (ES) | 0.53749174 |
| Normalized Enrichment Score (NES) | 1.9516042 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016938625 |
| FWER p-Value | 0.021 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0534 | Yes | ||
| 2 | PRKCH | 92 | 18.399 | 0.0788 | Yes | ||
| 3 | CA3 | 105 | 17.758 | 0.1068 | Yes | ||
| 4 | SATB1 | 140 | 15.729 | 0.1303 | Yes | ||
| 5 | SGK | 143 | 15.627 | 0.1554 | Yes | ||
| 6 | EDG5 | 146 | 15.468 | 0.1803 | Yes | ||
| 7 | CISH | 151 | 15.397 | 0.2049 | Yes | ||
| 8 | MEF2C | 185 | 14.237 | 0.2261 | Yes | ||
| 9 | HHEX | 190 | 14.145 | 0.2487 | Yes | ||
| 10 | BLNK | 239 | 13.319 | 0.2676 | Yes | ||
| 11 | BCL6B | 254 | 12.969 | 0.2878 | Yes | ||
| 12 | ERG | 426 | 10.702 | 0.2958 | Yes | ||
| 13 | LAT | 432 | 10.644 | 0.3127 | Yes | ||
| 14 | OPRM1 | 447 | 10.499 | 0.3288 | Yes | ||
| 15 | PDE4D | 524 | 9.718 | 0.3404 | Yes | ||
| 16 | PDE6D | 663 | 8.528 | 0.3467 | Yes | ||
| 17 | PPARGC1A | 665 | 8.507 | 0.3603 | Yes | ||
| 18 | MAP4K2 | 708 | 8.299 | 0.3715 | Yes | ||
| 19 | PDLIM2 | 729 | 8.184 | 0.3836 | Yes | ||
| 20 | HOXB4 | 768 | 7.921 | 0.3943 | Yes | ||
| 21 | STAT5A | 807 | 7.710 | 0.4047 | Yes | ||
| 22 | NFAT5 | 871 | 7.334 | 0.4131 | Yes | ||
| 23 | FEZ2 | 916 | 7.141 | 0.4222 | Yes | ||
| 24 | MBNL1 | 963 | 6.901 | 0.4309 | Yes | ||
| 25 | GNB2 | 1001 | 6.736 | 0.4397 | Yes | ||
| 26 | PRKCB1 | 1028 | 6.604 | 0.4490 | Yes | ||
| 27 | GPHN | 1070 | 6.470 | 0.4572 | Yes | ||
| 28 | CASKIN2 | 1085 | 6.396 | 0.4668 | Yes | ||
| 29 | HSD11B1 | 1105 | 6.325 | 0.4759 | Yes | ||
| 30 | TNFRSF19 | 1149 | 6.146 | 0.4835 | Yes | ||
| 31 | ZDHHC8 | 1217 | 5.849 | 0.4893 | Yes | ||
| 32 | IGF1 | 1238 | 5.779 | 0.4976 | Yes | ||
| 33 | RIN2 | 1296 | 5.538 | 0.5034 | Yes | ||
| 34 | SLC38A2 | 1421 | 5.175 | 0.5050 | Yes | ||
| 35 | LEPROTL1 | 1619 | 4.643 | 0.5019 | Yes | ||
| 36 | LIMK2 | 1627 | 4.618 | 0.5089 | Yes | ||
| 37 | RBMS1 | 1734 | 4.374 | 0.5102 | Yes | ||
| 38 | NOTCH4 | 1784 | 4.256 | 0.5144 | Yes | ||
| 39 | PITPNC1 | 1880 | 4.070 | 0.5159 | Yes | ||
| 40 | PBX1 | 1891 | 4.041 | 0.5218 | Yes | ||
| 41 | EGF | 1914 | 3.994 | 0.5271 | Yes | ||
| 42 | FEM1C | 1950 | 3.923 | 0.5315 | Yes | ||
| 43 | USP2 | 2010 | 3.801 | 0.5345 | Yes | ||
| 44 | TCF15 | 2065 | 3.695 | 0.5375 | Yes | ||
| 45 | MPO | 2256 | 3.396 | 0.5327 | No | ||
| 46 | TNKS1BP1 | 2282 | 3.333 | 0.5367 | No | ||
| 47 | TRPC4AP | 2381 | 3.164 | 0.5365 | No | ||
| 48 | SDCBP | 2505 | 3.002 | 0.5347 | No | ||
| 49 | LPL | 2667 | 2.767 | 0.5304 | No | ||
| 50 | CPA3 | 2751 | 2.683 | 0.5302 | No | ||
| 51 | LNX2 | 2942 | 2.455 | 0.5239 | No | ||
| 52 | ARPC2 | 3085 | 2.319 | 0.5199 | No | ||
| 53 | CSF3R | 3189 | 2.206 | 0.5179 | No | ||
| 54 | ARRDC3 | 3195 | 2.195 | 0.5212 | No | ||
| 55 | NCAM1 | 3634 | 1.827 | 0.5004 | No | ||
| 56 | ETS1 | 3675 | 1.798 | 0.5011 | No | ||
| 57 | FANK1 | 3721 | 1.769 | 0.5015 | No | ||
| 58 | PITX2 | 3728 | 1.764 | 0.5040 | No | ||
| 59 | RUNX2 | 4007 | 1.574 | 0.4915 | No | ||
| 60 | GRID2 | 4038 | 1.560 | 0.4924 | No | ||
| 61 | PHF15 | 4070 | 1.543 | 0.4932 | No | ||
| 62 | FN1 | 4076 | 1.540 | 0.4954 | No | ||
| 63 | LGI1 | 4404 | 1.348 | 0.4799 | No | ||
| 64 | SMOX | 4486 | 1.292 | 0.4776 | No | ||
| 65 | STAB1 | 4515 | 1.272 | 0.4781 | No | ||
| 66 | CTSK | 4538 | 1.258 | 0.4789 | No | ||
| 67 | NDFIP1 | 4541 | 1.255 | 0.4809 | No | ||
| 68 | CACNA1E | 4605 | 1.218 | 0.4794 | No | ||
| 69 | PTK7 | 4645 | 1.200 | 0.4792 | No | ||
| 70 | MITF | 4754 | 1.153 | 0.4752 | No | ||
| 71 | GRIA1 | 4759 | 1.150 | 0.4769 | No | ||
| 72 | JMJD1C | 4867 | 1.099 | 0.4728 | No | ||
| 73 | EML4 | 4975 | 1.048 | 0.4687 | No | ||
| 74 | DHH | 5073 | 1.004 | 0.4651 | No | ||
| 75 | VIT | 5114 | 0.986 | 0.4645 | No | ||
| 76 | MRPL24 | 5178 | 0.957 | 0.4626 | No | ||
| 77 | SLMAP | 5271 | 0.910 | 0.4591 | No | ||
| 78 | PHTF1 | 5345 | 0.879 | 0.4566 | No | ||
| 79 | PKP4 | 5380 | 0.866 | 0.4561 | No | ||
| 80 | NEXN | 5505 | 0.816 | 0.4507 | No | ||
| 81 | FGF12 | 5688 | 0.738 | 0.4421 | No | ||
| 82 | FLI1 | 5702 | 0.729 | 0.4425 | No | ||
| 83 | GIF | 5886 | 0.670 | 0.4337 | No | ||
| 84 | FGF10 | 5893 | 0.668 | 0.4344 | No | ||
| 85 | AP1G2 | 5994 | 0.628 | 0.4300 | No | ||
| 86 | NR4A1 | 6002 | 0.626 | 0.4307 | No | ||
| 87 | PAX8 | 6047 | 0.604 | 0.4293 | No | ||
| 88 | SHOX2 | 6354 | 0.486 | 0.4134 | No | ||
| 89 | MYL3 | 6459 | 0.449 | 0.4085 | No | ||
| 90 | CD3D | 6580 | 0.407 | 0.4027 | No | ||
| 91 | SLC36A2 | 6660 | 0.379 | 0.3990 | No | ||
| 92 | HOXC13 | 6670 | 0.376 | 0.3991 | No | ||
| 93 | RHOJ | 7159 | 0.216 | 0.3730 | No | ||
| 94 | IL21 | 7272 | 0.176 | 0.3672 | No | ||
| 95 | GSC | 7302 | 0.169 | 0.3659 | No | ||
| 96 | HNRPD | 7344 | 0.156 | 0.3640 | No | ||
| 97 | HOXD8 | 7554 | 0.099 | 0.3528 | No | ||
| 98 | CACNG3 | 8196 | -0.101 | 0.3182 | No | ||
| 99 | HOXD10 | 8430 | -0.170 | 0.3059 | No | ||
| 100 | CALR | 8545 | -0.206 | 0.3000 | No | ||
| 101 | ITGA8 | 8669 | -0.246 | 0.2937 | No | ||
| 102 | TMC3 | 8721 | -0.262 | 0.2914 | No | ||
| 103 | PHOX2B | 9299 | -0.428 | 0.2608 | No | ||
| 104 | POU4F1 | 9661 | -0.532 | 0.2421 | No | ||
| 105 | CRYGB | 9679 | -0.536 | 0.2420 | No | ||
| 106 | HOXB9 | 9688 | -0.538 | 0.2425 | No | ||
| 107 | RUNX3 | 9704 | -0.545 | 0.2425 | No | ||
| 108 | CITED2 | 9749 | -0.557 | 0.2410 | No | ||
| 109 | RHOQ | 9778 | -0.566 | 0.2404 | No | ||
| 110 | GREM1 | 9928 | -0.609 | 0.2333 | No | ||
| 111 | FGF8 | 9948 | -0.613 | 0.2333 | No | ||
| 112 | VSNL1 | 10030 | -0.639 | 0.2299 | No | ||
| 113 | STMN2 | 10067 | -0.648 | 0.2290 | No | ||
| 114 | CMYA5 | 10104 | -0.660 | 0.2282 | No | ||
| 115 | EPM2A | 10134 | -0.665 | 0.2277 | No | ||
| 116 | HIPK3 | 10456 | -0.753 | 0.2115 | No | ||
| 117 | SLC26A3 | 10840 | -0.859 | 0.1921 | No | ||
| 118 | A2BP1 | 10979 | -0.897 | 0.1861 | No | ||
| 119 | TLL1 | 11029 | -0.912 | 0.1849 | No | ||
| 120 | EVI1 | 11169 | -0.951 | 0.1789 | No | ||
| 121 | MAP2K7 | 11372 | -1.011 | 0.1696 | No | ||
| 122 | UBE2D3 | 11427 | -1.027 | 0.1683 | No | ||
| 123 | CSPG4 | 11676 | -1.098 | 0.1566 | No | ||
| 124 | HTR3B | 11811 | -1.141 | 0.1512 | No | ||
| 125 | ERF | 11926 | -1.175 | 0.1469 | No | ||
| 126 | TOM1L1 | 11941 | -1.179 | 0.1480 | No | ||
| 127 | FEV | 12009 | -1.199 | 0.1463 | No | ||
| 128 | GRIN2B | 12319 | -1.292 | 0.1317 | No | ||
| 129 | HOXC6 | 12350 | -1.302 | 0.1322 | No | ||
| 130 | BMP5 | 12366 | -1.309 | 0.1335 | No | ||
| 131 | PIK3C2G | 12451 | -1.335 | 0.1311 | No | ||
| 132 | PRCC | 12614 | -1.379 | 0.1245 | No | ||
| 133 | POU3F3 | 12677 | -1.403 | 0.1234 | No | ||
| 134 | NRG1 | 12758 | -1.429 | 0.1214 | No | ||
| 135 | TDRD9 | 12833 | -1.454 | 0.1197 | No | ||
| 136 | BDNF | 12898 | -1.477 | 0.1186 | No | ||
| 137 | TBC1D17 | 12939 | -1.489 | 0.1188 | No | ||
| 138 | ANGPTL1 | 13048 | -1.522 | 0.1154 | No | ||
| 139 | YARS | 13320 | -1.622 | 0.1034 | No | ||
| 140 | GPX1 | 13482 | -1.684 | 0.0974 | No | ||
| 141 | GPRC5D | 13605 | -1.731 | 0.0935 | No | ||
| 142 | FLT1 | 13723 | -1.780 | 0.0901 | No | ||
| 143 | UBL3 | 13759 | -1.793 | 0.0911 | No | ||
| 144 | OSR1 | 13803 | -1.813 | 0.0917 | No | ||
| 145 | SESTD1 | 13902 | -1.854 | 0.0893 | No | ||
| 146 | MAPK10 | 14173 | -1.978 | 0.0779 | No | ||
| 147 | HNF4G | 14405 | -2.093 | 0.0687 | No | ||
| 148 | DRD1IP | 14490 | -2.139 | 0.0676 | No | ||
| 149 | HSPB7 | 14534 | -2.165 | 0.0688 | No | ||
| 150 | NUDT4 | 14541 | -2.167 | 0.0720 | No | ||
| 151 | HOXB3 | 14553 | -2.175 | 0.0749 | No | ||
| 152 | EDN1 | 14566 | -2.181 | 0.0778 | No | ||
| 153 | AKT1S1 | 14898 | -2.361 | 0.0636 | No | ||
| 154 | DOCK4 | 15119 | -2.509 | 0.0557 | No | ||
| 155 | MTUS1 | 15353 | -2.685 | 0.0474 | No | ||
| 156 | BZW1 | 15377 | -2.699 | 0.0506 | No | ||
| 157 | ACBD6 | 15466 | -2.764 | 0.0502 | No | ||
| 158 | TTYH2 | 15774 | -3.055 | 0.0385 | No | ||
| 159 | CLOCK | 15937 | -3.233 | 0.0350 | No | ||
| 160 | SOSTDC1 | 15996 | -3.288 | 0.0371 | No | ||
| 161 | TRERF1 | 16095 | -3.410 | 0.0373 | No | ||
| 162 | CSRP3 | 16422 | -3.873 | 0.0259 | No | ||
| 163 | NEDD4 | 16589 | -4.146 | 0.0236 | No | ||
| 164 | TWIST1 | 16679 | -4.320 | 0.0257 | No | ||
| 165 | RNF138 | 16772 | -4.516 | 0.0280 | No | ||
| 166 | NRAS | 16794 | -4.560 | 0.0342 | No | ||
| 167 | CENTA1 | 16847 | -4.663 | 0.0389 | No | ||
| 168 | LRRC16 | 16949 | -4.841 | 0.0413 | No | ||
| 169 | POLR1D | 17100 | -5.209 | 0.0416 | No | ||
| 170 | EXTL2 | 18372 | -11.863 | -0.0082 | No | ||
| 171 | OSGEPL1 | 18465 | -13.252 | 0.0082 | No |