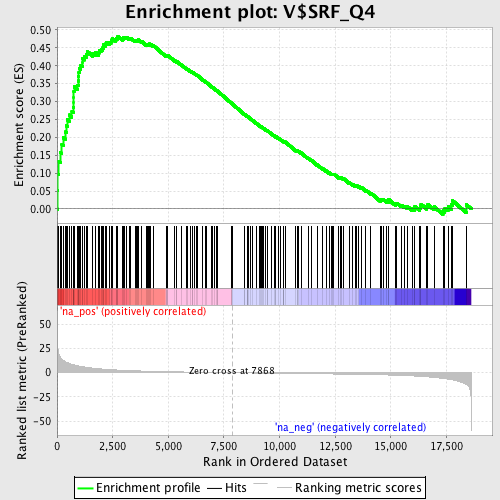
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SRF_Q4 |
| Enrichment Score (ES) | 0.48268557 |
| Normalized Enrichment Score (NES) | 1.760277 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.032350898 |
| FWER p-Value | 0.251 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABR | 21 | 28.176 | 0.0512 | Yes | ||
| 2 | EFHD1 | 32 | 25.080 | 0.0972 | Yes | ||
| 3 | MYLK | 71 | 20.288 | 0.1328 | Yes | ||
| 4 | SGK | 143 | 15.627 | 0.1580 | Yes | ||
| 5 | IHPK1 | 214 | 13.698 | 0.1796 | Yes | ||
| 6 | MAPKAPK2 | 276 | 12.560 | 0.1996 | Yes | ||
| 7 | SORT1 | 376 | 11.240 | 0.2152 | Yes | ||
| 8 | DUSP6 | 429 | 10.680 | 0.2322 | Yes | ||
| 9 | CSRP1 | 467 | 10.258 | 0.2492 | Yes | ||
| 10 | PPAP2B | 545 | 9.450 | 0.2626 | Yes | ||
| 11 | RASSF2 | 666 | 8.503 | 0.2719 | Yes | ||
| 12 | VCL | 736 | 8.130 | 0.2832 | Yes | ||
| 13 | SERPINH1 | 747 | 8.082 | 0.2977 | Yes | ||
| 14 | PPP1R12A | 750 | 8.075 | 0.3126 | Yes | ||
| 15 | MAPK14 | 754 | 8.054 | 0.3274 | Yes | ||
| 16 | HOXB4 | 768 | 7.921 | 0.3414 | Yes | ||
| 17 | EGR1 | 929 | 7.104 | 0.3459 | Yes | ||
| 18 | KRTCAP2 | 951 | 6.958 | 0.3577 | Yes | ||
| 19 | MBNL1 | 963 | 6.901 | 0.3699 | Yes | ||
| 20 | H2AFX | 978 | 6.835 | 0.3818 | Yes | ||
| 21 | DMD | 1013 | 6.671 | 0.3924 | Yes | ||
| 22 | HOXA5 | 1065 | 6.487 | 0.4016 | Yes | ||
| 23 | RSU1 | 1144 | 6.166 | 0.4089 | Yes | ||
| 24 | CHD2 | 1148 | 6.153 | 0.4201 | Yes | ||
| 25 | MCL1 | 1227 | 5.811 | 0.4267 | Yes | ||
| 26 | TLE3 | 1308 | 5.500 | 0.4326 | Yes | ||
| 27 | CDKN1B | 1354 | 5.343 | 0.4400 | Yes | ||
| 28 | TAZ | 1610 | 4.670 | 0.4349 | Yes | ||
| 29 | PLCB3 | 1732 | 4.377 | 0.4365 | Yes | ||
| 30 | PHF12 | 1877 | 4.077 | 0.4362 | Yes | ||
| 31 | MYL9 | 1892 | 4.040 | 0.4430 | Yes | ||
| 32 | MYL6 | 1978 | 3.868 | 0.4455 | Yes | ||
| 33 | ESRRA | 2031 | 3.765 | 0.4497 | Yes | ||
| 34 | DUSP2 | 2086 | 3.662 | 0.4536 | Yes | ||
| 35 | AKAP12 | 2090 | 3.653 | 0.4602 | Yes | ||
| 36 | MMP14 | 2194 | 3.492 | 0.4611 | Yes | ||
| 37 | TPM2 | 2230 | 3.447 | 0.4656 | Yes | ||
| 38 | NCOA6 | 2343 | 3.229 | 0.4655 | Yes | ||
| 39 | ROCK2 | 2424 | 3.107 | 0.4670 | Yes | ||
| 40 | ANXA6 | 2439 | 3.088 | 0.4719 | Yes | ||
| 41 | CFL2 | 2477 | 3.045 | 0.4756 | Yes | ||
| 42 | CALD1 | 2646 | 2.800 | 0.4717 | Yes | ||
| 43 | SVIL | 2649 | 2.798 | 0.4768 | Yes | ||
| 44 | TBL1X | 2700 | 2.733 | 0.4791 | Yes | ||
| 45 | ASPH | 2728 | 2.696 | 0.4827 | Yes | ||
| 46 | ERBB2IP | 2952 | 2.443 | 0.4751 | No | ||
| 47 | BLOC1S1 | 2972 | 2.426 | 0.4786 | No | ||
| 48 | BNC2 | 3029 | 2.366 | 0.4800 | No | ||
| 49 | CACNA1B | 3121 | 2.279 | 0.4793 | No | ||
| 50 | HOXB5 | 3241 | 2.156 | 0.4768 | No | ||
| 51 | DGKG | 3312 | 2.094 | 0.4769 | No | ||
| 52 | ELF4 | 3504 | 1.931 | 0.4701 | No | ||
| 53 | PPARG | 3566 | 1.890 | 0.4703 | No | ||
| 54 | EMILIN2 | 3628 | 1.835 | 0.4704 | No | ||
| 55 | SRF | 3642 | 1.823 | 0.4731 | No | ||
| 56 | COL23A1 | 3793 | 1.714 | 0.4682 | No | ||
| 57 | RERE | 4000 | 1.577 | 0.4599 | No | ||
| 58 | THRAP1 | 4083 | 1.533 | 0.4583 | No | ||
| 59 | MAP1A | 4110 | 1.522 | 0.4598 | No | ||
| 60 | CTGF | 4138 | 1.509 | 0.4611 | No | ||
| 61 | ELAVL4 | 4219 | 1.455 | 0.4595 | No | ||
| 62 | GAB2 | 4314 | 1.401 | 0.4570 | No | ||
| 63 | AGRP | 4910 | 1.080 | 0.4267 | No | ||
| 64 | ATRX | 4929 | 1.072 | 0.4277 | No | ||
| 65 | TADA3L | 4965 | 1.052 | 0.4278 | No | ||
| 66 | EML4 | 4975 | 1.048 | 0.4293 | No | ||
| 67 | FOXP1 | 5295 | 0.898 | 0.4136 | No | ||
| 68 | DIXDC1 | 5383 | 0.864 | 0.4105 | No | ||
| 69 | TAGLN | 5384 | 0.864 | 0.4121 | No | ||
| 70 | ASPA | 5596 | 0.776 | 0.4021 | No | ||
| 71 | MYH11 | 5803 | 0.698 | 0.3923 | No | ||
| 72 | MYO1B | 5881 | 0.671 | 0.3893 | No | ||
| 73 | NR4A1 | 6002 | 0.626 | 0.3840 | No | ||
| 74 | LPP | 6075 | 0.590 | 0.3812 | No | ||
| 75 | SRD5A2 | 6081 | 0.588 | 0.3820 | No | ||
| 76 | FOS | 6176 | 0.553 | 0.3779 | No | ||
| 77 | TGFB1I1 | 6278 | 0.514 | 0.3734 | No | ||
| 78 | MYL7 | 6317 | 0.497 | 0.3723 | No | ||
| 79 | SERTAD4 | 6555 | 0.417 | 0.3602 | No | ||
| 80 | HOXC5 | 6664 | 0.378 | 0.3550 | No | ||
| 81 | SDPR | 6709 | 0.361 | 0.3533 | No | ||
| 82 | FOSB | 6945 | 0.285 | 0.3411 | No | ||
| 83 | KCNMB1 | 7000 | 0.266 | 0.3387 | No | ||
| 84 | GPR20 | 7064 | 0.243 | 0.3357 | No | ||
| 85 | RHOJ | 7159 | 0.216 | 0.3310 | No | ||
| 86 | NKX2-2 | 7191 | 0.205 | 0.3297 | No | ||
| 87 | MRGPRF | 7217 | 0.195 | 0.3287 | No | ||
| 88 | SLC35A2 | 7817 | 0.013 | 0.2963 | No | ||
| 89 | TNNC1 | 7837 | 0.008 | 0.2953 | No | ||
| 90 | ARPC4 | 7885 | -0.005 | 0.2927 | No | ||
| 91 | SH3BGR | 8406 | -0.164 | 0.2649 | No | ||
| 92 | HOXD10 | 8430 | -0.170 | 0.2639 | No | ||
| 93 | MYO1E | 8535 | -0.203 | 0.2587 | No | ||
| 94 | DLL1 | 8567 | -0.212 | 0.2574 | No | ||
| 95 | EGR2 | 8574 | -0.213 | 0.2574 | No | ||
| 96 | STAT5B | 8605 | -0.224 | 0.2562 | No | ||
| 97 | MYL1 | 8697 | -0.256 | 0.2518 | No | ||
| 98 | FOXE3 | 8790 | -0.285 | 0.2473 | No | ||
| 99 | MUS81 | 8939 | -0.327 | 0.2399 | No | ||
| 100 | CX3CL1 | 9096 | -0.369 | 0.2321 | No | ||
| 101 | IER2 | 9158 | -0.386 | 0.2295 | No | ||
| 102 | HOXA3 | 9173 | -0.391 | 0.2295 | No | ||
| 103 | MSX1 | 9227 | -0.407 | 0.2274 | No | ||
| 104 | TNMD | 9291 | -0.427 | 0.2248 | No | ||
| 105 | RUNDC1 | 9387 | -0.454 | 0.2205 | No | ||
| 106 | DSTN | 9436 | -0.468 | 0.2187 | No | ||
| 107 | EGR4 | 9450 | -0.472 | 0.2189 | No | ||
| 108 | STAG2 | 9636 | -0.524 | 0.2098 | No | ||
| 109 | CITED2 | 9749 | -0.557 | 0.2048 | No | ||
| 110 | NPR3 | 9830 | -0.582 | 0.2016 | No | ||
| 111 | TMOD1 | 9833 | -0.583 | 0.2025 | No | ||
| 112 | FGF8 | 9948 | -0.613 | 0.1975 | No | ||
| 113 | FGF17 | 10027 | -0.638 | 0.1944 | No | ||
| 114 | SRP54 | 10169 | -0.674 | 0.1881 | No | ||
| 115 | JPH2 | 10177 | -0.676 | 0.1889 | No | ||
| 116 | THBS1 | 10193 | -0.682 | 0.1894 | No | ||
| 117 | FST | 10258 | -0.697 | 0.1872 | No | ||
| 118 | NLGN3 | 10729 | -0.833 | 0.1633 | No | ||
| 119 | ARHGEF17 | 10787 | -0.847 | 0.1618 | No | ||
| 120 | KCNK3 | 10802 | -0.849 | 0.1626 | No | ||
| 121 | MAP2K6 | 10855 | -0.863 | 0.1614 | No | ||
| 122 | FOXP2 | 10976 | -0.896 | 0.1565 | No | ||
| 123 | ACTG2 | 11278 | -0.980 | 0.1420 | No | ||
| 124 | IL17B | 11424 | -1.026 | 0.1361 | No | ||
| 125 | CRH | 11700 | -1.105 | 0.1232 | No | ||
| 126 | MID1 | 11932 | -1.176 | 0.1129 | No | ||
| 127 | ACTA1 | 12097 | -1.230 | 0.1063 | No | ||
| 128 | RIMS1 | 12226 | -1.266 | 0.1017 | No | ||
| 129 | RARB | 12347 | -1.301 | 0.0976 | No | ||
| 130 | RASD2 | 12398 | -1.317 | 0.0973 | No | ||
| 131 | FGFRL1 | 12440 | -1.329 | 0.0976 | No | ||
| 132 | TRIM46 | 12665 | -1.400 | 0.0880 | No | ||
| 133 | NPPA | 12739 | -1.424 | 0.0867 | No | ||
| 134 | GADD45G | 12780 | -1.438 | 0.0872 | No | ||
| 135 | PODN | 12879 | -1.471 | 0.0846 | No | ||
| 136 | ITGB1BP2 | 13144 | -1.556 | 0.0732 | No | ||
| 137 | DVL3 | 13281 | -1.604 | 0.0688 | No | ||
| 138 | CALN1 | 13402 | -1.652 | 0.0654 | No | ||
| 139 | SCOC | 13470 | -1.681 | 0.0649 | No | ||
| 140 | FLNA | 13554 | -1.716 | 0.0636 | No | ||
| 141 | JUNB | 13668 | -1.753 | 0.0607 | No | ||
| 142 | CKM | 13880 | -1.845 | 0.0527 | No | ||
| 143 | TANK | 14102 | -1.942 | 0.0443 | No | ||
| 144 | CNN1 | 14533 | -2.165 | 0.0250 | No | ||
| 145 | EDN1 | 14566 | -2.181 | 0.0273 | No | ||
| 146 | NR2F2 | 14654 | -2.224 | 0.0267 | No | ||
| 147 | ACTB | 14816 | -2.319 | 0.0223 | No | ||
| 148 | PFN1 | 14888 | -2.358 | 0.0229 | No | ||
| 149 | NPAS2 | 14890 | -2.359 | 0.0272 | No | ||
| 150 | SLC4A3 | 15209 | -2.573 | 0.0147 | No | ||
| 151 | ADAMTSL1 | 15272 | -2.620 | 0.0162 | No | ||
| 152 | LRRTM4 | 15470 | -2.766 | 0.0107 | No | ||
| 153 | GRK6 | 15616 | -2.892 | 0.0082 | No | ||
| 154 | INSM1 | 15737 | -3.003 | 0.0073 | No | ||
| 155 | VASP | 15953 | -3.246 | 0.0016 | No | ||
| 156 | TPM3 | 16051 | -3.357 | 0.0026 | No | ||
| 157 | NR2F1 | 16079 | -3.393 | 0.0075 | No | ||
| 158 | TAF5L | 16301 | -3.681 | 0.0023 | No | ||
| 159 | EGR3 | 16331 | -3.733 | 0.0077 | No | ||
| 160 | POU3F4 | 16353 | -3.758 | 0.0135 | No | ||
| 161 | TPM1 | 16613 | -4.188 | 0.0072 | No | ||
| 162 | KCNA1 | 16665 | -4.283 | 0.0124 | No | ||
| 163 | CFL1 | 16942 | -4.834 | 0.0064 | No | ||
| 164 | RBBP7 | 17347 | -5.853 | -0.0046 | No | ||
| 165 | PADI2 | 17431 | -6.092 | 0.0022 | No | ||
| 166 | EEF1B2 | 17575 | -6.609 | 0.0068 | No | ||
| 167 | NPM3 | 17721 | -7.173 | 0.0122 | No | ||
| 168 | SLC25A4 | 17766 | -7.356 | 0.0235 | No | ||
| 169 | CSDA | 18392 | -12.136 | 0.0121 | No |