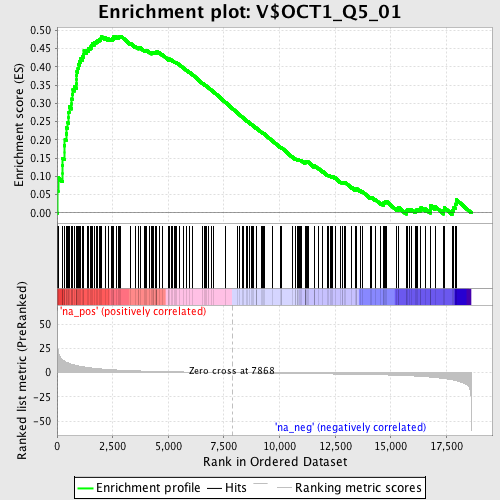
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$OCT1_Q5_01 |
| Enrichment Score (ES) | 0.4843263 |
| Normalized Enrichment Score (NES) | 1.7813202 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.049760815 |
| FWER p-Value | 0.199 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STAT4 | 7 | 37.571 | 0.0607 | Yes | ||
| 2 | HOXA7 | 43 | 23.284 | 0.0966 | Yes | ||
| 3 | BLNK | 239 | 13.319 | 0.1077 | Yes | ||
| 4 | GPRC5B | 242 | 13.296 | 0.1291 | Yes | ||
| 5 | VPREB3 | 250 | 13.198 | 0.1502 | Yes | ||
| 6 | AXUD1 | 332 | 11.882 | 0.1651 | Yes | ||
| 7 | CYR61 | 333 | 11.874 | 0.1844 | Yes | ||
| 8 | TSGA14 | 353 | 11.617 | 0.2023 | Yes | ||
| 9 | TCEAL1 | 407 | 10.843 | 0.2170 | Yes | ||
| 10 | DUSP6 | 429 | 10.680 | 0.2332 | Yes | ||
| 11 | SLC25A12 | 480 | 10.118 | 0.2469 | Yes | ||
| 12 | SCML4 | 518 | 9.779 | 0.2608 | Yes | ||
| 13 | PDE4D | 524 | 9.718 | 0.2763 | Yes | ||
| 14 | ATF7IP | 541 | 9.500 | 0.2909 | Yes | ||
| 15 | THRA | 657 | 8.562 | 0.2986 | Yes | ||
| 16 | DLX1 | 662 | 8.535 | 0.3122 | Yes | ||
| 17 | THRAP5 | 680 | 8.447 | 0.3250 | Yes | ||
| 18 | RHOB | 710 | 8.295 | 0.3369 | Yes | ||
| 19 | HOXB4 | 768 | 7.921 | 0.3467 | Yes | ||
| 20 | RDH11 | 866 | 7.376 | 0.3534 | Yes | ||
| 21 | RRAS | 873 | 7.329 | 0.3650 | Yes | ||
| 22 | MSI2 | 878 | 7.286 | 0.3766 | Yes | ||
| 23 | PCYT1B | 887 | 7.239 | 0.3880 | Yes | ||
| 24 | H3F3B | 935 | 7.045 | 0.3969 | Yes | ||
| 25 | ARHGAP4 | 967 | 6.888 | 0.4064 | Yes | ||
| 26 | DMD | 1013 | 6.671 | 0.4148 | Yes | ||
| 27 | HOXA5 | 1065 | 6.487 | 0.4225 | Yes | ||
| 28 | FCHSD1 | 1146 | 6.160 | 0.4282 | Yes | ||
| 29 | SH3BGRL | 1197 | 5.931 | 0.4351 | Yes | ||
| 30 | SH3GL3 | 1207 | 5.894 | 0.4442 | Yes | ||
| 31 | DLGAP4 | 1343 | 5.387 | 0.4456 | Yes | ||
| 32 | BTK | 1419 | 5.180 | 0.4500 | Yes | ||
| 33 | NPHP4 | 1483 | 5.000 | 0.4547 | Yes | ||
| 34 | HIST1H2BM | 1546 | 4.831 | 0.4592 | Yes | ||
| 35 | DPYSL2 | 1581 | 4.741 | 0.4650 | Yes | ||
| 36 | TPD52 | 1679 | 4.495 | 0.4671 | Yes | ||
| 37 | PTEN | 1750 | 4.330 | 0.4703 | Yes | ||
| 38 | KIF13A | 1830 | 4.176 | 0.4728 | Yes | ||
| 39 | SREBF2 | 1886 | 4.052 | 0.4764 | Yes | ||
| 40 | HIST1H3B | 1969 | 3.888 | 0.4783 | Yes | ||
| 41 | HIST1H2BK | 1981 | 3.860 | 0.4840 | Yes | ||
| 42 | ETV1 | 2167 | 3.528 | 0.4797 | Yes | ||
| 43 | BAT2 | 2318 | 3.271 | 0.4768 | Yes | ||
| 44 | EYA1 | 2422 | 3.111 | 0.4763 | Yes | ||
| 45 | HIST1H2AB | 2484 | 3.032 | 0.4779 | Yes | ||
| 46 | SFRS14 | 2525 | 2.974 | 0.4806 | Yes | ||
| 47 | NFYB | 2550 | 2.924 | 0.4840 | Yes | ||
| 48 | LPL | 2667 | 2.767 | 0.4822 | Yes | ||
| 49 | ADORA2A | 2767 | 2.664 | 0.4812 | Yes | ||
| 50 | NFIA | 2793 | 2.635 | 0.4841 | Yes | ||
| 51 | ALK | 2866 | 2.538 | 0.4843 | Yes | ||
| 52 | TBXAS1 | 3296 | 2.108 | 0.4645 | No | ||
| 53 | HIST1H2BB | 3507 | 1.929 | 0.4562 | No | ||
| 54 | SRF | 3642 | 1.823 | 0.4519 | No | ||
| 55 | RAB26 | 3659 | 1.808 | 0.4540 | No | ||
| 56 | DPYSL3 | 3734 | 1.759 | 0.4528 | No | ||
| 57 | MAB21L2 | 3948 | 1.611 | 0.4439 | No | ||
| 58 | GRIA3 | 3970 | 1.598 | 0.4453 | No | ||
| 59 | ANXA2 | 4008 | 1.574 | 0.4459 | No | ||
| 60 | TLL2 | 4153 | 1.500 | 0.4405 | No | ||
| 61 | GNA14 | 4251 | 1.435 | 0.4376 | No | ||
| 62 | SILV | 4274 | 1.426 | 0.4387 | No | ||
| 63 | GAB2 | 4314 | 1.401 | 0.4389 | No | ||
| 64 | NXPH1 | 4341 | 1.389 | 0.4397 | No | ||
| 65 | LRRN1 | 4420 | 1.340 | 0.4377 | No | ||
| 66 | ADORA1 | 4457 | 1.308 | 0.4378 | No | ||
| 67 | SPRR2B | 4462 | 1.306 | 0.4397 | No | ||
| 68 | HIST1H3C | 4482 | 1.296 | 0.4408 | No | ||
| 69 | PIGT | 4483 | 1.296 | 0.4429 | No | ||
| 70 | PLA1A | 4585 | 1.228 | 0.4394 | No | ||
| 71 | HIST1H2AH | 4730 | 1.167 | 0.4335 | No | ||
| 72 | ARMET | 5009 | 1.033 | 0.4201 | No | ||
| 73 | HIST1H2BC | 5014 | 1.031 | 0.4216 | No | ||
| 74 | HIST1H2AC | 5039 | 1.018 | 0.4219 | No | ||
| 75 | UPK3A | 5119 | 0.982 | 0.4192 | No | ||
| 76 | CRISP1 | 5199 | 0.946 | 0.4165 | No | ||
| 77 | FOXP1 | 5295 | 0.898 | 0.4128 | No | ||
| 78 | HIST2H2AC | 5333 | 0.884 | 0.4122 | No | ||
| 79 | HOXB8 | 5376 | 0.867 | 0.4113 | No | ||
| 80 | TCF12 | 5491 | 0.821 | 0.4065 | No | ||
| 81 | FGF12 | 5688 | 0.738 | 0.3971 | No | ||
| 82 | CNNM4 | 5826 | 0.692 | 0.3907 | No | ||
| 83 | ACCN1 | 5961 | 0.642 | 0.3845 | No | ||
| 84 | FZD4 | 6080 | 0.588 | 0.3791 | No | ||
| 85 | SERTAD4 | 6555 | 0.417 | 0.3540 | No | ||
| 86 | LYN | 6633 | 0.389 | 0.3505 | No | ||
| 87 | HOXC5 | 6664 | 0.378 | 0.3495 | No | ||
| 88 | HOXB6 | 6733 | 0.354 | 0.3464 | No | ||
| 89 | NOS1 | 6809 | 0.328 | 0.3428 | No | ||
| 90 | PCDH11X | 6942 | 0.286 | 0.3361 | No | ||
| 91 | NIN | 7027 | 0.258 | 0.3320 | No | ||
| 92 | HOXD8 | 7554 | 0.099 | 0.3036 | No | ||
| 93 | GPR4 | 8094 | -0.073 | 0.2745 | No | ||
| 94 | ITSN1 | 8188 | -0.099 | 0.2696 | No | ||
| 95 | ARF6 | 8328 | -0.141 | 0.2623 | No | ||
| 96 | MRAS | 8375 | -0.155 | 0.2601 | No | ||
| 97 | SEMA6C | 8534 | -0.202 | 0.2518 | No | ||
| 98 | EGR2 | 8574 | -0.213 | 0.2500 | No | ||
| 99 | PFKFB1 | 8644 | -0.237 | 0.2467 | No | ||
| 100 | SLC6A15 | 8735 | -0.265 | 0.2422 | No | ||
| 101 | EHF | 8765 | -0.276 | 0.2411 | No | ||
| 102 | TCERG1L | 8838 | -0.297 | 0.2377 | No | ||
| 103 | OTX2 | 8943 | -0.328 | 0.2326 | No | ||
| 104 | SFRP1 | 8948 | -0.330 | 0.2329 | No | ||
| 105 | ALDH1A1 | 8973 | -0.335 | 0.2321 | No | ||
| 106 | HIST1H2AA | 9182 | -0.393 | 0.2215 | No | ||
| 107 | SYNPR | 9236 | -0.410 | 0.2193 | No | ||
| 108 | NRL | 9270 | -0.422 | 0.2182 | No | ||
| 109 | PHOX2B | 9299 | -0.428 | 0.2173 | No | ||
| 110 | STX12 | 9689 | -0.539 | 0.1971 | No | ||
| 111 | VSNL1 | 10030 | -0.639 | 0.1797 | No | ||
| 112 | POU2F3 | 10081 | -0.654 | 0.1781 | No | ||
| 113 | PRICKLE2 | 10091 | -0.657 | 0.1786 | No | ||
| 114 | NDNL2 | 10578 | -0.787 | 0.1536 | No | ||
| 115 | NEUROG1 | 10707 | -0.826 | 0.1479 | No | ||
| 116 | LIPG | 10736 | -0.835 | 0.1478 | No | ||
| 117 | NRXN3 | 10804 | -0.850 | 0.1455 | No | ||
| 118 | RPP21 | 10815 | -0.852 | 0.1464 | No | ||
| 119 | MAP2K6 | 10855 | -0.863 | 0.1457 | No | ||
| 120 | ATP2A2 | 10896 | -0.876 | 0.1449 | No | ||
| 121 | TRAF3 | 10940 | -0.887 | 0.1440 | No | ||
| 122 | FOXP2 | 10976 | -0.896 | 0.1436 | No | ||
| 123 | SMPX | 11143 | -0.943 | 0.1361 | No | ||
| 124 | PAX6 | 11145 | -0.944 | 0.1376 | No | ||
| 125 | CDX4 | 11152 | -0.945 | 0.1388 | No | ||
| 126 | FGF14 | 11163 | -0.949 | 0.1398 | No | ||
| 127 | ASCL3 | 11184 | -0.952 | 0.1403 | No | ||
| 128 | PRRX1 | 11202 | -0.958 | 0.1409 | No | ||
| 129 | FSTL5 | 11251 | -0.971 | 0.1399 | No | ||
| 130 | CRYZL1 | 11281 | -0.982 | 0.1399 | No | ||
| 131 | LHX6 | 11566 | -1.064 | 0.1262 | No | ||
| 132 | REL | 11573 | -1.065 | 0.1276 | No | ||
| 133 | TCF2 | 11574 | -1.065 | 0.1293 | No | ||
| 134 | HOXD11 | 11743 | -1.121 | 0.1221 | No | ||
| 135 | MID1 | 11932 | -1.176 | 0.1138 | No | ||
| 136 | PTHLH | 12160 | -1.248 | 0.1035 | No | ||
| 137 | LRCH4 | 12204 | -1.261 | 0.1032 | No | ||
| 138 | IRX4 | 12282 | -1.282 | 0.1011 | No | ||
| 139 | RARB | 12347 | -1.301 | 0.0997 | No | ||
| 140 | PPP2R5C | 12397 | -1.317 | 0.0992 | No | ||
| 141 | SLITRK6 | 12493 | -1.348 | 0.0963 | No | ||
| 142 | MGAT5B | 12748 | -1.426 | 0.0848 | No | ||
| 143 | FBXO24 | 12805 | -1.445 | 0.0841 | No | ||
| 144 | HPCAL4 | 12837 | -1.457 | 0.0848 | No | ||
| 145 | BDNF | 12898 | -1.477 | 0.0839 | No | ||
| 146 | FGF20 | 12944 | -1.491 | 0.0839 | No | ||
| 147 | CSF3 | 13246 | -1.592 | 0.0702 | No | ||
| 148 | SP6 | 13411 | -1.656 | 0.0640 | No | ||
| 149 | TMOD2 | 13441 | -1.670 | 0.0651 | No | ||
| 150 | SCOC | 13470 | -1.681 | 0.0663 | No | ||
| 151 | PHF6 | 13615 | -1.735 | 0.0613 | No | ||
| 152 | UBE2S | 13718 | -1.777 | 0.0587 | No | ||
| 153 | WNT6 | 14071 | -1.930 | 0.0427 | No | ||
| 154 | CLCA3 | 14145 | -1.965 | 0.0419 | No | ||
| 155 | GPM6A | 14309 | -2.048 | 0.0364 | No | ||
| 156 | HOXB3 | 14553 | -2.175 | 0.0268 | No | ||
| 157 | PCDH8 | 14666 | -2.234 | 0.0243 | No | ||
| 158 | E2F3 | 14675 | -2.239 | 0.0275 | No | ||
| 159 | HOXA10 | 14722 | -2.262 | 0.0287 | No | ||
| 160 | GPR85 | 14752 | -2.278 | 0.0308 | No | ||
| 161 | COL25A1 | 14800 | -2.311 | 0.0320 | No | ||
| 162 | DAPK3 | 15265 | -2.613 | 0.0111 | No | ||
| 163 | HIST1H2BA | 15330 | -2.666 | 0.0120 | No | ||
| 164 | MTUS1 | 15353 | -2.685 | 0.0151 | No | ||
| 165 | PRDM1 | 15689 | -2.948 | 0.0018 | No | ||
| 166 | SFRP2 | 15706 | -2.965 | 0.0057 | No | ||
| 167 | BCL2 | 15743 | -3.007 | 0.0086 | No | ||
| 168 | CDX2 | 15835 | -3.128 | 0.0088 | No | ||
| 169 | ZHX2 | 15934 | -3.231 | 0.0087 | No | ||
| 170 | TRERF1 | 16095 | -3.410 | 0.0056 | No | ||
| 171 | ISL1 | 16148 | -3.468 | 0.0084 | No | ||
| 172 | RTTN | 16219 | -3.576 | 0.0104 | No | ||
| 173 | CDK2 | 16319 | -3.710 | 0.0111 | No | ||
| 174 | POU3F4 | 16353 | -3.758 | 0.0154 | No | ||
| 175 | EAF2 | 16553 | -4.083 | 0.0112 | No | ||
| 176 | GALNT1 | 16767 | -4.500 | 0.0070 | No | ||
| 177 | CD86 | 16785 | -4.549 | 0.0134 | No | ||
| 178 | NRAS | 16794 | -4.560 | 0.0204 | No | ||
| 179 | NRXN1 | 16987 | -4.906 | 0.0180 | No | ||
| 180 | DSCR1 | 17374 | -5.932 | 0.0067 | No | ||
| 181 | KCNN3 | 17396 | -6.000 | 0.0153 | No | ||
| 182 | SFRS2 | 17755 | -7.315 | 0.0077 | No | ||
| 183 | LDB2 | 17832 | -7.657 | 0.0161 | No | ||
| 184 | PIM2 | 17916 | -8.044 | 0.0246 | No | ||
| 185 | TSNAX | 17939 | -8.187 | 0.0367 | No |