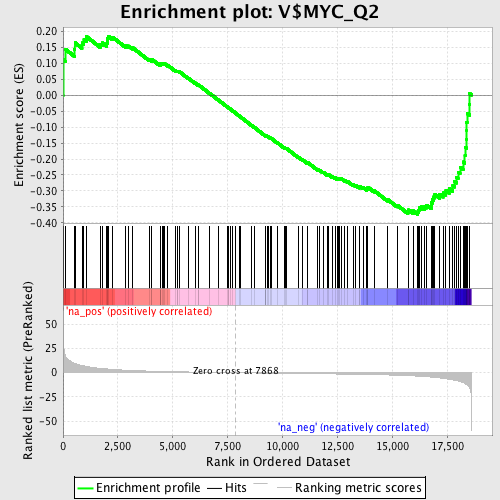
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | -0.3735522 |
| Normalized Enrichment Score (NES) | -1.3274161 |
| Nominal p-value | 0.05487805 |
| FDR q-value | 0.37687394 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FXYD6 | 3 | 48.657 | 0.1107 | No | ||
| 2 | BCL9L | 121 | 16.738 | 0.1425 | No | ||
| 3 | PSEN2 | 521 | 9.760 | 0.1431 | No | ||
| 4 | ATF7IP | 541 | 9.500 | 0.1638 | No | ||
| 5 | PFKFB3 | 861 | 7.407 | 0.1634 | No | ||
| 6 | KRTCAP2 | 951 | 6.958 | 0.1744 | No | ||
| 7 | CDC14A | 1063 | 6.501 | 0.1832 | No | ||
| 8 | TMEM24 | 1688 | 4.469 | 0.1597 | No | ||
| 9 | FCHSD2 | 1797 | 4.240 | 0.1635 | No | ||
| 10 | USP15 | 1993 | 3.836 | 0.1617 | No | ||
| 11 | USP2 | 2010 | 3.801 | 0.1695 | No | ||
| 12 | ESRRA | 2031 | 3.765 | 0.1770 | No | ||
| 13 | CHD4 | 2049 | 3.732 | 0.1846 | No | ||
| 14 | COMMD3 | 2249 | 3.403 | 0.1816 | No | ||
| 15 | RAB3IL1 | 2865 | 2.539 | 0.1541 | No | ||
| 16 | GPD1 | 2975 | 2.420 | 0.1537 | No | ||
| 17 | H3F3A | 3157 | 2.238 | 0.1490 | No | ||
| 18 | ALDH3B1 | 3931 | 1.618 | 0.1109 | No | ||
| 19 | RPL22 | 4036 | 1.561 | 0.1089 | No | ||
| 20 | NIT1 | 4050 | 1.556 | 0.1117 | No | ||
| 21 | PTPRF | 4425 | 1.335 | 0.0945 | No | ||
| 22 | SC5DL | 4436 | 1.329 | 0.0970 | No | ||
| 23 | COL2A1 | 4450 | 1.316 | 0.0993 | No | ||
| 24 | CBX5 | 4545 | 1.254 | 0.0971 | No | ||
| 25 | AP3M1 | 4587 | 1.228 | 0.0977 | No | ||
| 26 | CTBP2 | 4617 | 1.215 | 0.0989 | No | ||
| 27 | SLC1A7 | 4771 | 1.146 | 0.0932 | No | ||
| 28 | RRAGC | 5122 | 0.980 | 0.0765 | No | ||
| 29 | LTBR | 5191 | 0.951 | 0.0750 | No | ||
| 30 | SCYL1 | 5231 | 0.928 | 0.0750 | No | ||
| 31 | HMGN2 | 5302 | 0.896 | 0.0733 | No | ||
| 32 | PITX3 | 5715 | 0.725 | 0.0526 | No | ||
| 33 | LIN28 | 6013 | 0.622 | 0.0380 | No | ||
| 34 | HPCA | 6152 | 0.562 | 0.0318 | No | ||
| 35 | CNNM1 | 6180 | 0.549 | 0.0316 | No | ||
| 36 | HOXC13 | 6670 | 0.376 | 0.0060 | No | ||
| 37 | NRIP3 | 7083 | 0.239 | -0.0157 | No | ||
| 38 | FOSL1 | 7483 | 0.116 | -0.0370 | No | ||
| 39 | SIRT1 | 7525 | 0.105 | -0.0390 | No | ||
| 40 | FOXD3 | 7552 | 0.099 | -0.0402 | No | ||
| 41 | UBXD3 | 7623 | 0.075 | -0.0438 | No | ||
| 42 | AGMAT | 7736 | 0.037 | -0.0497 | No | ||
| 43 | APOA5 | 7860 | 0.003 | -0.0564 | No | ||
| 44 | FADS3 | 8044 | -0.058 | -0.0661 | No | ||
| 45 | B4GALT2 | 8087 | -0.072 | -0.0682 | No | ||
| 46 | FABP3 | 8599 | -0.222 | -0.0954 | No | ||
| 47 | SLC6A15 | 8735 | -0.265 | -0.1021 | No | ||
| 48 | B3GALT2 | 9229 | -0.407 | -0.1278 | No | ||
| 49 | SLC16A1 | 9234 | -0.408 | -0.1271 | No | ||
| 50 | LMX1A | 9239 | -0.411 | -0.1263 | No | ||
| 51 | RUSC1 | 9318 | -0.434 | -0.1296 | No | ||
| 52 | HNRPF | 9375 | -0.450 | -0.1316 | No | ||
| 53 | LHX9 | 9437 | -0.469 | -0.1338 | No | ||
| 54 | CTSF | 9497 | -0.485 | -0.1359 | No | ||
| 55 | CENTB5 | 9759 | -0.560 | -0.1487 | No | ||
| 56 | RORC | 10073 | -0.650 | -0.1642 | No | ||
| 57 | ILK | 10128 | -0.664 | -0.1656 | No | ||
| 58 | TAGLN2 | 10181 | -0.678 | -0.1668 | No | ||
| 59 | TAF6L | 10720 | -0.831 | -0.1940 | No | ||
| 60 | SYT6 | 10917 | -0.882 | -0.2026 | No | ||
| 61 | WEE1 | 11125 | -0.938 | -0.2117 | No | ||
| 62 | PAX6 | 11145 | -0.944 | -0.2105 | No | ||
| 63 | STMN1 | 11601 | -1.074 | -0.2327 | No | ||
| 64 | NKX2-3 | 11681 | -1.100 | -0.2345 | No | ||
| 65 | IPO7 | 11849 | -1.155 | -0.2409 | No | ||
| 66 | HNRPA1 | 12031 | -1.207 | -0.2479 | No | ||
| 67 | TDRD1 | 12074 | -1.220 | -0.2474 | No | ||
| 68 | RFX5 | 12274 | -1.280 | -0.2552 | No | ||
| 69 | CHRM1 | 12393 | -1.317 | -0.2586 | No | ||
| 70 | IPO13 | 12508 | -1.351 | -0.2617 | No | ||
| 71 | LRP8 | 12534 | -1.356 | -0.2600 | No | ||
| 72 | NR0B2 | 12605 | -1.376 | -0.2606 | No | ||
| 73 | TRIM46 | 12665 | -1.400 | -0.2606 | No | ||
| 74 | HPCAL4 | 12837 | -1.457 | -0.2665 | No | ||
| 75 | OPRD1 | 12963 | -1.499 | -0.2699 | No | ||
| 76 | CGN | 13227 | -1.584 | -0.2805 | No | ||
| 77 | TESK2 | 13331 | -1.627 | -0.2823 | No | ||
| 78 | BHLHB3 | 13491 | -1.689 | -0.2871 | No | ||
| 79 | PLA2G4A | 13526 | -1.703 | -0.2850 | No | ||
| 80 | ALX4 | 13675 | -1.758 | -0.2890 | No | ||
| 81 | POU3F1 | 13847 | -1.831 | -0.2941 | No | ||
| 82 | POGK | 13879 | -1.845 | -0.2916 | No | ||
| 83 | HOXC12 | 13890 | -1.848 | -0.2879 | No | ||
| 84 | IL15RA | 14179 | -1.979 | -0.2990 | No | ||
| 85 | FXYD2 | 14784 | -2.302 | -0.3264 | No | ||
| 86 | RTN4RL2 | 15225 | -2.586 | -0.3443 | No | ||
| 87 | TIAL1 | 15720 | -2.984 | -0.3642 | No | ||
| 88 | TGFB2 | 15757 | -3.017 | -0.3593 | No | ||
| 89 | AMPD2 | 15949 | -3.243 | -0.3622 | No | ||
| 90 | GTF2H1 | 16160 | -3.494 | -0.3656 | Yes | ||
| 91 | ATP6V0B | 16200 | -3.544 | -0.3596 | Yes | ||
| 92 | ZZZ3 | 16226 | -3.586 | -0.3528 | Yes | ||
| 93 | ATP5F1 | 16323 | -3.717 | -0.3495 | Yes | ||
| 94 | EIF3S10 | 16482 | -3.975 | -0.3490 | Yes | ||
| 95 | TFB2M | 16564 | -4.101 | -0.3441 | Yes | ||
| 96 | PFDN2 | 16784 | -4.545 | -0.3455 | Yes | ||
| 97 | NRAS | 16794 | -4.560 | -0.3356 | Yes | ||
| 98 | EIF4B | 16838 | -4.646 | -0.3274 | Yes | ||
| 99 | RHEBL1 | 16878 | -4.718 | -0.3187 | Yes | ||
| 100 | CFL1 | 16942 | -4.834 | -0.3111 | Yes | ||
| 101 | B3GALT6 | 17163 | -5.379 | -0.3108 | Yes | ||
| 102 | PA2G4 | 17316 | -5.750 | -0.3059 | Yes | ||
| 103 | SFXN2 | 17448 | -6.163 | -0.2989 | Yes | ||
| 104 | ARL3 | 17593 | -6.686 | -0.2915 | Yes | ||
| 105 | PABPC4 | 17764 | -7.353 | -0.2839 | Yes | ||
| 106 | PPRC1 | 17835 | -7.677 | -0.2702 | Yes | ||
| 107 | TIMM10 | 17928 | -8.102 | -0.2567 | Yes | ||
| 108 | IVNS1ABP | 18004 | -8.562 | -0.2413 | Yes | ||
| 109 | COPS7A | 18093 | -9.164 | -0.2252 | Yes | ||
| 110 | NUDC | 18231 | -10.291 | -0.2091 | Yes | ||
| 111 | ATAD3A | 18316 | -11.079 | -0.1885 | Yes | ||
| 112 | EBNA1BP2 | 18326 | -11.235 | -0.1633 | Yes | ||
| 113 | AK2 | 18378 | -12.015 | -0.1387 | Yes | ||
| 114 | SHMT2 | 18386 | -12.073 | -0.1116 | Yes | ||
| 115 | CSDA | 18392 | -12.136 | -0.0842 | Yes | ||
| 116 | LDHA | 18415 | -12.382 | -0.0572 | Yes | ||
| 117 | FKBP11 | 18519 | -14.634 | -0.0295 | Yes | ||
| 118 | ADK | 18531 | -15.211 | 0.0046 | Yes |
