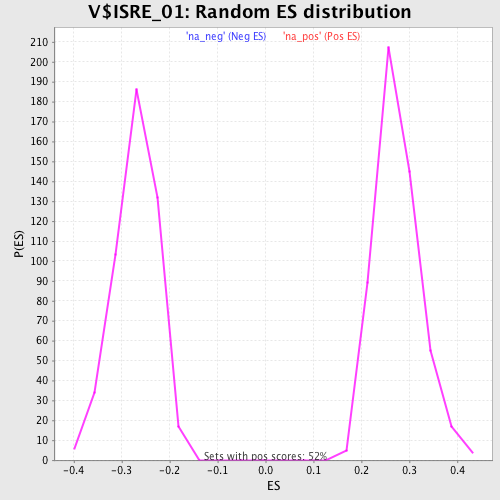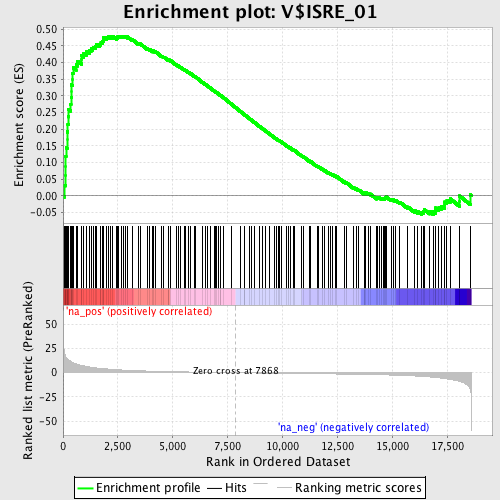
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$ISRE_01 |
| Enrichment Score (ES) | 0.47892714 |
| Normalized Enrichment Score (NES) | 1.7409953 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03744619 |
| FWER p-Value | 0.305 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCF7L2 | 72 | 20.268 | 0.0316 | Yes | ||
| 2 | ESR1 | 115 | 17.309 | 0.0597 | Yes | ||
| 3 | UBE1L | 119 | 16.848 | 0.0890 | Yes | ||
| 4 | SSBP2 | 122 | 16.680 | 0.1181 | Yes | ||
| 5 | USP18 | 135 | 15.860 | 0.1453 | Yes | ||
| 6 | MEF2C | 185 | 14.237 | 0.1675 | Yes | ||
| 7 | GNB4 | 187 | 14.219 | 0.1924 | Yes | ||
| 8 | ISGF3G | 216 | 13.694 | 0.2149 | Yes | ||
| 9 | BLNK | 239 | 13.319 | 0.2370 | Yes | ||
| 10 | IFIT2 | 253 | 13.006 | 0.2591 | Yes | ||
| 11 | CD151 | 349 | 11.654 | 0.2744 | Yes | ||
| 12 | BCL11A | 362 | 11.533 | 0.2939 | Yes | ||
| 13 | MAP3K11 | 373 | 11.326 | 0.3132 | Yes | ||
| 14 | ETV6 | 375 | 11.290 | 0.3329 | Yes | ||
| 15 | ERG | 426 | 10.702 | 0.3490 | Yes | ||
| 16 | SERHL | 430 | 10.660 | 0.3675 | Yes | ||
| 17 | MEIS1 | 458 | 10.375 | 0.3842 | Yes | ||
| 18 | SLC12A7 | 592 | 9.063 | 0.3929 | Yes | ||
| 19 | PPARGC1A | 665 | 8.507 | 0.4039 | Yes | ||
| 20 | TCIRG1 | 839 | 7.526 | 0.4077 | Yes | ||
| 21 | CDKN2C | 845 | 7.476 | 0.4205 | Yes | ||
| 22 | ETV5 | 941 | 7.015 | 0.4277 | Yes | ||
| 23 | BBX | 1051 | 6.537 | 0.4332 | Yes | ||
| 24 | SH3BGRL | 1197 | 5.931 | 0.4358 | Yes | ||
| 25 | DGKA | 1275 | 5.622 | 0.4414 | Yes | ||
| 26 | MYB | 1362 | 5.320 | 0.4461 | Yes | ||
| 27 | B2M | 1468 | 5.057 | 0.4493 | Yes | ||
| 28 | GSDMDC1 | 1517 | 4.897 | 0.4553 | Yes | ||
| 29 | MOV10 | 1681 | 4.484 | 0.4543 | Yes | ||
| 30 | ZFPM1 | 1724 | 4.397 | 0.4597 | Yes | ||
| 31 | TLR7 | 1793 | 4.247 | 0.4635 | Yes | ||
| 32 | IRF2 | 1831 | 4.176 | 0.4688 | Yes | ||
| 33 | DDX58 | 1837 | 4.157 | 0.4758 | Yes | ||
| 34 | CHKA | 1988 | 3.847 | 0.4744 | Yes | ||
| 35 | TCF15 | 2065 | 3.695 | 0.4768 | Yes | ||
| 36 | PML | 2156 | 3.538 | 0.4781 | Yes | ||
| 37 | DAPP1 | 2260 | 3.382 | 0.4784 | Yes | ||
| 38 | ADAM15 | 2453 | 3.075 | 0.4734 | Yes | ||
| 39 | CDK6 | 2494 | 3.022 | 0.4766 | Yes | ||
| 40 | RRBP1 | 2546 | 2.928 | 0.4789 | Yes | ||
| 41 | FYCO1 | 2645 | 2.801 | 0.4785 | No | ||
| 42 | PLAG1 | 2740 | 2.688 | 0.4781 | No | ||
| 43 | CXCR4 | 2830 | 2.592 | 0.4779 | No | ||
| 44 | BST2 | 2923 | 2.478 | 0.4772 | No | ||
| 45 | HPCAL1 | 3161 | 2.233 | 0.4683 | No | ||
| 46 | ZNF385 | 3433 | 1.981 | 0.4571 | No | ||
| 47 | OPTN | 3521 | 1.921 | 0.4558 | No | ||
| 48 | TOP1 | 3843 | 1.682 | 0.4413 | No | ||
| 49 | MMP25 | 3951 | 1.609 | 0.4383 | No | ||
| 50 | THRAP1 | 4083 | 1.533 | 0.4339 | No | ||
| 51 | PLXNC1 | 4115 | 1.520 | 0.4349 | No | ||
| 52 | UNC5C | 4199 | 1.468 | 0.4330 | No | ||
| 53 | GPR18 | 4506 | 1.277 | 0.4187 | No | ||
| 54 | PCDH7 | 4595 | 1.223 | 0.4160 | No | ||
| 55 | PROK2 | 4782 | 1.139 | 0.4080 | No | ||
| 56 | JMJD2A | 4816 | 1.122 | 0.4081 | No | ||
| 57 | ZBP1 | 4903 | 1.083 | 0.4054 | No | ||
| 58 | IL22RA1 | 5162 | 0.965 | 0.3931 | No | ||
| 59 | AIF1 | 5248 | 0.921 | 0.3901 | No | ||
| 60 | SLC11A2 | 5362 | 0.873 | 0.3855 | No | ||
| 61 | GPSN2 | 5517 | 0.811 | 0.3786 | No | ||
| 62 | ASPA | 5596 | 0.776 | 0.3757 | No | ||
| 63 | PITX3 | 5715 | 0.725 | 0.3706 | No | ||
| 64 | LRP2 | 5817 | 0.694 | 0.3664 | No | ||
| 65 | IFNB1 | 5987 | 0.632 | 0.3583 | No | ||
| 66 | OTX1 | 6014 | 0.621 | 0.3580 | No | ||
| 67 | SLC24A1 | 6358 | 0.485 | 0.3403 | No | ||
| 68 | EREG | 6496 | 0.437 | 0.3336 | No | ||
| 69 | PRDM16 | 6578 | 0.408 | 0.3299 | No | ||
| 70 | SDPR | 6709 | 0.361 | 0.3235 | No | ||
| 71 | EYA4 | 6907 | 0.295 | 0.3134 | No | ||
| 72 | HIST1H1C | 6943 | 0.286 | 0.3120 | No | ||
| 73 | TNFSF15 | 6989 | 0.270 | 0.3100 | No | ||
| 74 | KYNU | 7062 | 0.244 | 0.3065 | No | ||
| 75 | SIX1 | 7177 | 0.210 | 0.3007 | No | ||
| 76 | NKX2-2 | 7191 | 0.205 | 0.3004 | No | ||
| 77 | OSBP | 7294 | 0.171 | 0.2952 | No | ||
| 78 | HTR2C | 7682 | 0.055 | 0.2743 | No | ||
| 79 | IL27 | 8095 | -0.074 | 0.2521 | No | ||
| 80 | SLA | 8271 | -0.125 | 0.2429 | No | ||
| 81 | CRY1 | 8284 | -0.127 | 0.2424 | No | ||
| 82 | IL4 | 8484 | -0.187 | 0.2320 | No | ||
| 83 | EGR2 | 8574 | -0.213 | 0.2275 | No | ||
| 84 | CXCL10 | 8733 | -0.265 | 0.2194 | No | ||
| 85 | PURA | 8934 | -0.326 | 0.2092 | No | ||
| 86 | LMO1 | 9100 | -0.370 | 0.2009 | No | ||
| 87 | MSX1 | 9227 | -0.407 | 0.1948 | No | ||
| 88 | DUSP10 | 9386 | -0.454 | 0.1870 | No | ||
| 89 | CDH3 | 9643 | -0.525 | 0.1741 | No | ||
| 90 | TBX21 | 9726 | -0.550 | 0.1706 | No | ||
| 91 | NPR3 | 9830 | -0.582 | 0.1660 | No | ||
| 92 | ZCCHC5 | 9840 | -0.584 | 0.1666 | No | ||
| 93 | FMR1 | 9937 | -0.610 | 0.1625 | No | ||
| 94 | THBS1 | 10193 | -0.682 | 0.1498 | No | ||
| 95 | FIGN | 10265 | -0.699 | 0.1472 | No | ||
| 96 | EPN1 | 10346 | -0.723 | 0.1442 | No | ||
| 97 | JARID2 | 10491 | -0.764 | 0.1377 | No | ||
| 98 | PRKACA | 10529 | -0.773 | 0.1370 | No | ||
| 99 | MAP2K6 | 10855 | -0.863 | 0.1210 | No | ||
| 100 | MET | 10966 | -0.894 | 0.1166 | No | ||
| 101 | CACNA2D2 | 11218 | -0.963 | 0.1047 | No | ||
| 102 | LSP1 | 11287 | -0.984 | 0.1027 | No | ||
| 103 | COL12A1 | 11571 | -1.065 | 0.0892 | No | ||
| 104 | FGF9 | 11632 | -1.082 | 0.0879 | No | ||
| 105 | WNT8B | 11807 | -1.140 | 0.0805 | No | ||
| 106 | KCNJ16 | 11923 | -1.173 | 0.0763 | No | ||
| 107 | TAPBP | 12086 | -1.225 | 0.0697 | No | ||
| 108 | PSMA3 | 12197 | -1.259 | 0.0659 | No | ||
| 109 | MIA2 | 12281 | -1.282 | 0.0637 | No | ||
| 110 | PPP2R5C | 12397 | -1.317 | 0.0597 | No | ||
| 111 | CXCL11 | 12471 | -1.340 | 0.0581 | No | ||
| 112 | SLC25A28 | 12817 | -1.449 | 0.0420 | No | ||
| 113 | BDNF | 12898 | -1.477 | 0.0402 | No | ||
| 114 | EGFL6 | 13250 | -1.593 | 0.0240 | No | ||
| 115 | CBX4 | 13351 | -1.635 | 0.0215 | No | ||
| 116 | GPX1 | 13482 | -1.684 | 0.0174 | No | ||
| 117 | LGP2 | 13744 | -1.787 | 0.0064 | No | ||
| 118 | GATA6 | 13755 | -1.791 | 0.0090 | No | ||
| 119 | PAX2 | 13775 | -1.802 | 0.0111 | No | ||
| 120 | KCNE4 | 13925 | -1.865 | 0.0063 | No | ||
| 121 | TNFRSF17 | 14003 | -1.902 | 0.0055 | No | ||
| 122 | TNFSF13B | 14286 | -2.029 | -0.0063 | No | ||
| 123 | SEMA6D | 14315 | -2.050 | -0.0042 | No | ||
| 124 | CXXC5 | 14409 | -2.096 | -0.0055 | No | ||
| 125 | KPNA3 | 14520 | -2.158 | -0.0077 | No | ||
| 126 | ERBB2 | 14583 | -2.186 | -0.0072 | No | ||
| 127 | OVOL1 | 14653 | -2.224 | -0.0071 | No | ||
| 128 | E2F3 | 14675 | -2.239 | -0.0043 | No | ||
| 129 | PSMA5 | 14721 | -2.261 | -0.0028 | No | ||
| 130 | USF1 | 14954 | -2.401 | -0.0111 | No | ||
| 131 | TNRC4 | 15038 | -2.453 | -0.0113 | No | ||
| 132 | PIGR | 15146 | -2.529 | -0.0127 | No | ||
| 133 | STAT6 | 15352 | -2.683 | -0.0191 | No | ||
| 134 | PGK1 | 15715 | -2.975 | -0.0335 | No | ||
| 135 | RANGAP1 | 16032 | -3.337 | -0.0448 | No | ||
| 136 | MS4A1 | 16171 | -3.503 | -0.0461 | No | ||
| 137 | NT5C3 | 16356 | -3.765 | -0.0495 | No | ||
| 138 | RIPK2 | 16429 | -3.880 | -0.0466 | No | ||
| 139 | AMMECR1 | 16455 | -3.927 | -0.0410 | No | ||
| 140 | ADD3 | 16707 | -4.375 | -0.0470 | No | ||
| 141 | CRK | 16873 | -4.709 | -0.0477 | No | ||
| 142 | YWHAG | 16965 | -4.874 | -0.0440 | No | ||
| 143 | ANAPC1 | 16982 | -4.901 | -0.0363 | No | ||
| 144 | GPR65 | 17129 | -5.290 | -0.0350 | No | ||
| 145 | PIK3R3 | 17231 | -5.558 | -0.0307 | No | ||
| 146 | HEPH | 17388 | -5.978 | -0.0287 | No | ||
| 147 | KCNN3 | 17396 | -6.000 | -0.0185 | No | ||
| 148 | ARHGEF6 | 17493 | -6.346 | -0.0126 | No | ||
| 149 | EHD4 | 17639 | -6.858 | -0.0085 | No | ||
| 150 | LY86 | 18065 | -8.964 | -0.0158 | No | ||
| 151 | HOMER1 | 18071 | -9.004 | -0.0003 | No | ||
| 152 | ANGPTL4 | 18556 | -16.972 | 0.0032 | No |