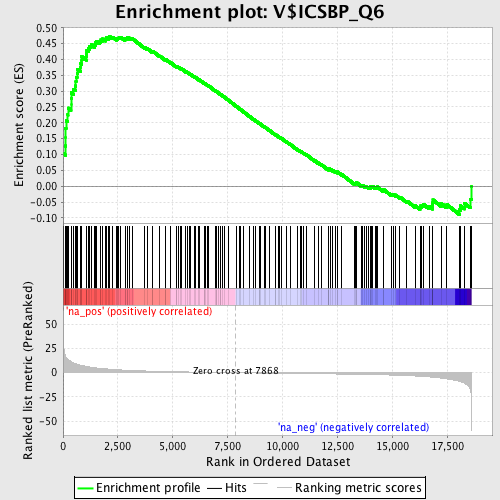
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$ICSBP_Q6 |
| Enrichment Score (ES) | 0.47357652 |
| Normalized Enrichment Score (NES) | 1.7342839 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03624244 |
| FWER p-Value | 0.327 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 63.431 | 0.1034 | Yes | ||
| 2 | ATP10A | 102 | 17.971 | 0.1272 | Yes | ||
| 3 | ESR1 | 115 | 17.309 | 0.1547 | Yes | ||
| 4 | UBE1L | 119 | 16.848 | 0.1820 | Yes | ||
| 5 | USP18 | 135 | 15.860 | 0.2070 | Yes | ||
| 6 | ADAR | 215 | 13.696 | 0.2251 | Yes | ||
| 7 | IL18BP | 223 | 13.597 | 0.2469 | Yes | ||
| 8 | VGLL4 | 360 | 11.542 | 0.2583 | Yes | ||
| 9 | BCL11A | 362 | 11.533 | 0.2770 | Yes | ||
| 10 | MAP3K11 | 373 | 11.326 | 0.2949 | Yes | ||
| 11 | FUT8 | 477 | 10.142 | 0.3059 | Yes | ||
| 12 | FES | 563 | 9.289 | 0.3164 | Yes | ||
| 13 | CASP7 | 584 | 9.126 | 0.3302 | Yes | ||
| 14 | SLC12A7 | 592 | 9.063 | 0.3446 | Yes | ||
| 15 | LGALS8 | 636 | 8.700 | 0.3564 | Yes | ||
| 16 | PPARGC1A | 665 | 8.507 | 0.3688 | Yes | ||
| 17 | PSMB9 | 795 | 7.768 | 0.3745 | Yes | ||
| 18 | TAP1 | 806 | 7.714 | 0.3865 | Yes | ||
| 19 | TCIRG1 | 839 | 7.526 | 0.3970 | Yes | ||
| 20 | CDKN2C | 845 | 7.476 | 0.4089 | Yes | ||
| 21 | BBX | 1051 | 6.537 | 0.4085 | Yes | ||
| 22 | LRMP | 1062 | 6.503 | 0.4185 | Yes | ||
| 23 | PRDX5 | 1079 | 6.427 | 0.4281 | Yes | ||
| 24 | TNFRSF19 | 1149 | 6.146 | 0.4344 | Yes | ||
| 25 | ITPR1 | 1193 | 5.947 | 0.4418 | Yes | ||
| 26 | DGKA | 1275 | 5.622 | 0.4465 | Yes | ||
| 27 | HOXB7 | 1443 | 5.104 | 0.4458 | Yes | ||
| 28 | B2M | 1468 | 5.057 | 0.4527 | Yes | ||
| 29 | GSDMDC1 | 1517 | 4.897 | 0.4581 | Yes | ||
| 30 | MOV10 | 1681 | 4.484 | 0.4566 | Yes | ||
| 31 | ZFPM1 | 1724 | 4.397 | 0.4615 | Yes | ||
| 32 | TLR7 | 1793 | 4.247 | 0.4647 | Yes | ||
| 33 | PSMB8 | 1917 | 3.986 | 0.4646 | Yes | ||
| 34 | ELK4 | 1954 | 3.914 | 0.4690 | Yes | ||
| 35 | TCF15 | 2065 | 3.695 | 0.4690 | Yes | ||
| 36 | ITPR3 | 2092 | 3.648 | 0.4736 | Yes | ||
| 37 | DAPP1 | 2260 | 3.382 | 0.4700 | No | ||
| 38 | DDB2 | 2429 | 3.104 | 0.4660 | No | ||
| 39 | CDK6 | 2494 | 3.022 | 0.4675 | No | ||
| 40 | RRBP1 | 2546 | 2.928 | 0.4695 | No | ||
| 41 | NFKB1 | 2617 | 2.841 | 0.4703 | No | ||
| 42 | DIO2 | 2821 | 2.606 | 0.4635 | No | ||
| 43 | CXCR4 | 2830 | 2.592 | 0.4673 | No | ||
| 44 | MGAT4A | 2914 | 2.487 | 0.4669 | No | ||
| 45 | BST2 | 2923 | 2.478 | 0.4705 | No | ||
| 46 | CSAD | 3046 | 2.350 | 0.4677 | No | ||
| 47 | HPCAL1 | 3161 | 2.233 | 0.4652 | No | ||
| 48 | CABLES1 | 3715 | 1.773 | 0.4381 | No | ||
| 49 | TOP1 | 3843 | 1.682 | 0.4340 | No | ||
| 50 | PHF15 | 4070 | 1.543 | 0.4242 | No | ||
| 51 | THRAP1 | 4083 | 1.533 | 0.4261 | No | ||
| 52 | LGI1 | 4404 | 1.348 | 0.4109 | No | ||
| 53 | SOS1 | 4644 | 1.201 | 0.4000 | No | ||
| 54 | HIVEP1 | 4662 | 1.194 | 0.4010 | No | ||
| 55 | ZBP1 | 4903 | 1.083 | 0.3897 | No | ||
| 56 | IL22RA1 | 5162 | 0.965 | 0.3773 | No | ||
| 57 | TFEC | 5173 | 0.961 | 0.3784 | No | ||
| 58 | AIF1 | 5248 | 0.921 | 0.3759 | No | ||
| 59 | SLC11A2 | 5362 | 0.873 | 0.3712 | No | ||
| 60 | PKP4 | 5380 | 0.866 | 0.3716 | No | ||
| 61 | ASPA | 5596 | 0.776 | 0.3613 | No | ||
| 62 | TBX1 | 5648 | 0.754 | 0.3597 | No | ||
| 63 | ADAM8 | 5752 | 0.713 | 0.3553 | No | ||
| 64 | LRP2 | 5817 | 0.694 | 0.3530 | No | ||
| 65 | IFNB1 | 5987 | 0.632 | 0.3448 | No | ||
| 66 | OTX1 | 6014 | 0.621 | 0.3444 | No | ||
| 67 | COL4A2 | 6191 | 0.547 | 0.3358 | No | ||
| 68 | SLC15A3 | 6195 | 0.546 | 0.3365 | No | ||
| 69 | ATP11C | 6421 | 0.464 | 0.3251 | No | ||
| 70 | RCOR1 | 6436 | 0.458 | 0.3251 | No | ||
| 71 | EREG | 6496 | 0.437 | 0.3226 | No | ||
| 72 | PRDM16 | 6578 | 0.408 | 0.3189 | No | ||
| 73 | PAX3 | 6621 | 0.393 | 0.3172 | No | ||
| 74 | HIST1H1C | 6943 | 0.286 | 0.3003 | No | ||
| 75 | TNFSF15 | 6989 | 0.270 | 0.2983 | No | ||
| 76 | CHST1 | 7008 | 0.264 | 0.2978 | No | ||
| 77 | KYNU | 7062 | 0.244 | 0.2953 | No | ||
| 78 | SIX1 | 7177 | 0.210 | 0.2895 | No | ||
| 79 | NKX2-2 | 7191 | 0.205 | 0.2891 | No | ||
| 80 | IL21 | 7272 | 0.176 | 0.2850 | No | ||
| 81 | HNRPD | 7344 | 0.156 | 0.2815 | No | ||
| 82 | NPAS3 | 7548 | 0.100 | 0.2706 | No | ||
| 83 | ENPP2 | 7917 | -0.013 | 0.2507 | No | ||
| 84 | USP28 | 8027 | -0.052 | 0.2449 | No | ||
| 85 | IL27 | 8095 | -0.074 | 0.2414 | No | ||
| 86 | COL4A1 | 8239 | -0.115 | 0.2338 | No | ||
| 87 | GZMK | 8489 | -0.191 | 0.2206 | No | ||
| 88 | ASCL2 | 8694 | -0.255 | 0.2100 | No | ||
| 89 | HAS3 | 8752 | -0.272 | 0.2074 | No | ||
| 90 | PURA | 8934 | -0.326 | 0.1981 | No | ||
| 91 | ZIC5 | 8945 | -0.329 | 0.1981 | No | ||
| 92 | ALDH1A1 | 8973 | -0.335 | 0.1972 | No | ||
| 93 | HOXA3 | 9173 | -0.391 | 0.1870 | No | ||
| 94 | MSX1 | 9227 | -0.407 | 0.1848 | No | ||
| 95 | DUSP10 | 9386 | -0.454 | 0.1770 | No | ||
| 96 | POU4F1 | 9661 | -0.532 | 0.1630 | No | ||
| 97 | ECM2 | 9692 | -0.541 | 0.1623 | No | ||
| 98 | NPR3 | 9830 | -0.582 | 0.1558 | No | ||
| 99 | CXCL16 | 9884 | -0.597 | 0.1539 | No | ||
| 100 | FMR1 | 9937 | -0.610 | 0.1521 | No | ||
| 101 | THBS1 | 10193 | -0.682 | 0.1394 | No | ||
| 102 | EPN1 | 10346 | -0.723 | 0.1323 | No | ||
| 103 | ENTPD1 | 10687 | -0.821 | 0.1152 | No | ||
| 104 | HAPLN1 | 10814 | -0.852 | 0.1098 | No | ||
| 105 | MAP2K6 | 10855 | -0.863 | 0.1090 | No | ||
| 106 | TEK | 10972 | -0.896 | 0.1042 | No | ||
| 107 | KCNJ1 | 11087 | -0.928 | 0.0995 | No | ||
| 108 | SATB2 | 11470 | -1.036 | 0.0805 | No | ||
| 109 | FGF9 | 11632 | -1.082 | 0.0736 | No | ||
| 110 | CYP7A1 | 11760 | -1.126 | 0.0685 | No | ||
| 111 | TAPBP | 12086 | -1.225 | 0.0529 | No | ||
| 112 | HOXA13 | 12089 | -1.226 | 0.0548 | No | ||
| 113 | KLHL13 | 12098 | -1.230 | 0.0564 | No | ||
| 114 | PSMA3 | 12197 | -1.259 | 0.0531 | No | ||
| 115 | CNTFR | 12298 | -1.286 | 0.0498 | No | ||
| 116 | PPP2R5C | 12397 | -1.317 | 0.0466 | No | ||
| 117 | SLITRK6 | 12493 | -1.348 | 0.0437 | No | ||
| 118 | ATP5H | 12498 | -1.348 | 0.0457 | No | ||
| 119 | LRRN3 | 12698 | -1.409 | 0.0372 | No | ||
| 120 | ELAVL2 | 13287 | -1.609 | 0.0079 | No | ||
| 121 | MPDZ | 13310 | -1.619 | 0.0094 | No | ||
| 122 | CBX4 | 13351 | -1.635 | 0.0099 | No | ||
| 123 | INDO | 13372 | -1.642 | 0.0115 | No | ||
| 124 | PHF6 | 13615 | -1.735 | 0.0012 | No | ||
| 125 | IFI35 | 13648 | -1.744 | 0.0023 | No | ||
| 126 | GATA6 | 13755 | -1.791 | -0.0005 | No | ||
| 127 | LY75 | 13839 | -1.828 | -0.0020 | No | ||
| 128 | LAMA3 | 13926 | -1.865 | -0.0037 | No | ||
| 129 | TNFRSF17 | 14003 | -1.902 | -0.0047 | No | ||
| 130 | BANK1 | 14008 | -1.903 | -0.0018 | No | ||
| 131 | SP8 | 14068 | -1.927 | -0.0019 | No | ||
| 132 | TRPM3 | 14085 | -1.934 | 0.0004 | No | ||
| 133 | GPR22 | 14223 | -1.998 | -0.0037 | No | ||
| 134 | TNFSF13B | 14286 | -2.029 | -0.0038 | No | ||
| 135 | SEMA6D | 14315 | -2.050 | -0.0020 | No | ||
| 136 | ERBB2 | 14583 | -2.186 | -0.0129 | No | ||
| 137 | IRAK1 | 14586 | -2.187 | -0.0094 | No | ||
| 138 | USF1 | 14954 | -2.401 | -0.0254 | No | ||
| 139 | TNRC4 | 15038 | -2.453 | -0.0259 | No | ||
| 140 | PIGR | 15146 | -2.529 | -0.0276 | No | ||
| 141 | STAT6 | 15352 | -2.683 | -0.0343 | No | ||
| 142 | CTSS | 15670 | -2.931 | -0.0467 | No | ||
| 143 | MXI1 | 16041 | -3.347 | -0.0613 | No | ||
| 144 | OSR2 | 16268 | -3.641 | -0.0676 | No | ||
| 145 | ZBTB33 | 16279 | -3.653 | -0.0622 | No | ||
| 146 | NT5C3 | 16356 | -3.765 | -0.0602 | No | ||
| 147 | RIPK2 | 16429 | -3.880 | -0.0578 | No | ||
| 148 | TWIST1 | 16679 | -4.320 | -0.0642 | No | ||
| 149 | SP110 | 16826 | -4.629 | -0.0646 | No | ||
| 150 | SOCS1 | 16831 | -4.638 | -0.0572 | No | ||
| 151 | NDRG3 | 16851 | -4.675 | -0.0506 | No | ||
| 152 | SYNCRIP | 16857 | -4.692 | -0.0433 | No | ||
| 153 | PIK3R3 | 17231 | -5.558 | -0.0544 | No | ||
| 154 | ARHGEF6 | 17493 | -6.346 | -0.0582 | No | ||
| 155 | LY86 | 18065 | -8.964 | -0.0745 | No | ||
| 156 | EIF5A | 18110 | -9.263 | -0.0618 | No | ||
| 157 | CASK | 18296 | -10.829 | -0.0542 | No | ||
| 158 | ANGPTL4 | 18556 | -16.972 | -0.0406 | No | ||
| 159 | PTK2 | 18609 | -26.874 | 0.0004 | No |