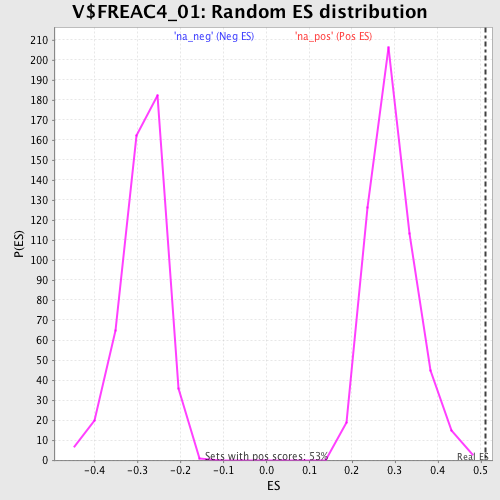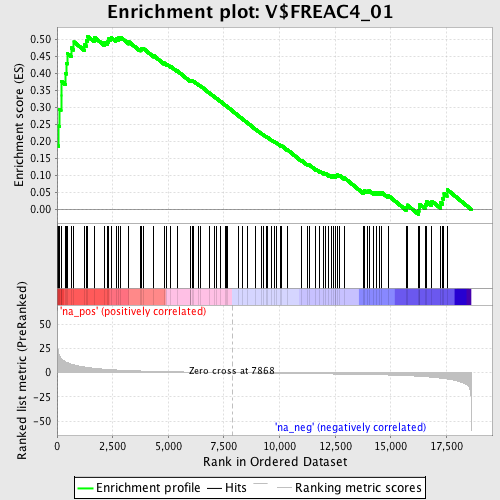
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FREAC4_01 |
| Enrichment Score (ES) | 0.5100782 |
| Normalized Enrichment Score (NES) | 1.7386619 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.036607858 |
| FWER p-Value | 0.312 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 63.431 | 0.1891 | Yes | ||
| 2 | TCF7L2 | 72 | 20.268 | 0.2457 | Yes | ||
| 3 | CDKN1A | 113 | 17.385 | 0.2954 | Yes | ||
| 4 | LEF1 | 176 | 14.378 | 0.3349 | Yes | ||
| 5 | GFRA1 | 177 | 14.371 | 0.3778 | Yes | ||
| 6 | BTBD3 | 370 | 11.391 | 0.4014 | Yes | ||
| 7 | ERG | 426 | 10.702 | 0.4303 | Yes | ||
| 8 | MBNL2 | 470 | 10.212 | 0.4585 | Yes | ||
| 9 | ETHE1 | 635 | 8.710 | 0.4756 | Yes | ||
| 10 | CTCF | 758 | 8.038 | 0.4930 | Yes | ||
| 11 | IGF1 | 1238 | 5.779 | 0.4843 | Yes | ||
| 12 | TLE3 | 1308 | 5.500 | 0.4970 | Yes | ||
| 13 | DUSP1 | 1361 | 5.322 | 0.5101 | Yes | ||
| 14 | CAMK1D | 1666 | 4.519 | 0.5071 | No | ||
| 15 | PELI2 | 2139 | 3.557 | 0.4923 | No | ||
| 16 | PER2 | 2279 | 3.339 | 0.4947 | No | ||
| 17 | PLS3 | 2326 | 3.263 | 0.5020 | No | ||
| 18 | EYA1 | 2422 | 3.111 | 0.5061 | No | ||
| 19 | CALD1 | 2646 | 2.800 | 0.5024 | No | ||
| 20 | ULK1 | 2749 | 2.684 | 0.5049 | No | ||
| 21 | KBTBD2 | 2858 | 2.547 | 0.5067 | No | ||
| 22 | ZADH2 | 3227 | 2.164 | 0.4933 | No | ||
| 23 | PITX2 | 3728 | 1.764 | 0.4715 | No | ||
| 24 | TCF1 | 3777 | 1.726 | 0.4741 | No | ||
| 25 | UBE2H | 3862 | 1.664 | 0.4745 | No | ||
| 26 | SLC24A2 | 4345 | 1.382 | 0.4526 | No | ||
| 27 | IBSP | 4804 | 1.128 | 0.4312 | No | ||
| 28 | MYH2 | 4937 | 1.067 | 0.4273 | No | ||
| 29 | VIT | 5114 | 0.986 | 0.4207 | No | ||
| 30 | ZFX | 5391 | 0.861 | 0.4084 | No | ||
| 31 | AP1G2 | 5994 | 0.628 | 0.3778 | No | ||
| 32 | NR4A1 | 6002 | 0.626 | 0.3793 | No | ||
| 33 | OTX1 | 6014 | 0.621 | 0.3805 | No | ||
| 34 | HOXA11 | 6086 | 0.585 | 0.3784 | No | ||
| 35 | PNRC1 | 6147 | 0.564 | 0.3769 | No | ||
| 36 | SHOX2 | 6354 | 0.486 | 0.3672 | No | ||
| 37 | PDCD1LG2 | 6461 | 0.449 | 0.3628 | No | ||
| 38 | OTOP3 | 6865 | 0.309 | 0.3420 | No | ||
| 39 | IER5 | 7073 | 0.241 | 0.3315 | No | ||
| 40 | PTHR1 | 7176 | 0.210 | 0.3266 | No | ||
| 41 | GABRR1 | 7342 | 0.156 | 0.3182 | No | ||
| 42 | FOXD3 | 7552 | 0.099 | 0.3072 | No | ||
| 43 | UBXD3 | 7623 | 0.075 | 0.3037 | No | ||
| 44 | PHEX | 7656 | 0.062 | 0.3021 | No | ||
| 45 | SYT10 | 8149 | -0.088 | 0.2758 | No | ||
| 46 | PLUNC | 8326 | -0.141 | 0.2667 | No | ||
| 47 | ROR1 | 8577 | -0.214 | 0.2539 | No | ||
| 48 | PURA | 8934 | -0.326 | 0.2356 | No | ||
| 49 | SIN3A | 9176 | -0.392 | 0.2238 | No | ||
| 50 | TNMD | 9291 | -0.427 | 0.2189 | No | ||
| 51 | LOX | 9407 | -0.459 | 0.2141 | No | ||
| 52 | HOXD9 | 9456 | -0.474 | 0.2129 | No | ||
| 53 | TGFB3 | 9645 | -0.526 | 0.2043 | No | ||
| 54 | CITED2 | 9749 | -0.557 | 0.2004 | No | ||
| 55 | ADRB1 | 9865 | -0.590 | 0.1960 | No | ||
| 56 | SLC34A3 | 10051 | -0.644 | 0.1879 | No | ||
| 57 | SPRY4 | 10071 | -0.649 | 0.1888 | No | ||
| 58 | PUM2 | 10342 | -0.722 | 0.1764 | No | ||
| 59 | FOXP2 | 10976 | -0.896 | 0.1449 | No | ||
| 60 | PHTF2 | 11243 | -0.969 | 0.1334 | No | ||
| 61 | MSC | 11340 | -1.000 | 0.1312 | No | ||
| 62 | FGF9 | 11632 | -1.082 | 0.1187 | No | ||
| 63 | TLX1 | 11805 | -1.139 | 0.1128 | No | ||
| 64 | HSPG2 | 11966 | -1.190 | 0.1077 | No | ||
| 65 | FERD3L | 12044 | -1.210 | 0.1072 | No | ||
| 66 | MCC | 12218 | -1.264 | 0.1016 | No | ||
| 67 | HOXC6 | 12350 | -1.302 | 0.0984 | No | ||
| 68 | EVX1 | 12401 | -1.319 | 0.0997 | No | ||
| 69 | KLF12 | 12519 | -1.353 | 0.0974 | No | ||
| 70 | TECTA | 12522 | -1.354 | 0.1013 | No | ||
| 71 | CD109 | 12587 | -1.370 | 0.1019 | No | ||
| 72 | POU3F3 | 12677 | -1.403 | 0.1013 | No | ||
| 73 | COLEC12 | 12914 | -1.483 | 0.0930 | No | ||
| 74 | ALDH1A2 | 13783 | -1.806 | 0.0515 | No | ||
| 75 | RARG | 13800 | -1.812 | 0.0561 | No | ||
| 76 | FOXB1 | 13933 | -1.869 | 0.0545 | No | ||
| 77 | NNAT | 14033 | -1.914 | 0.0549 | No | ||
| 78 | ODZ1 | 14225 | -2.000 | 0.0505 | No | ||
| 79 | TRPS1 | 14365 | -2.071 | 0.0492 | No | ||
| 80 | HR | 14469 | -2.130 | 0.0500 | No | ||
| 81 | CLDN18 | 14590 | -2.189 | 0.0500 | No | ||
| 82 | TBX4 | 14874 | -2.350 | 0.0418 | No | ||
| 83 | PRDM1 | 15689 | -2.948 | 0.0066 | No | ||
| 84 | SLC31A1 | 15732 | -2.998 | 0.0133 | No | ||
| 85 | NCDN | 16251 | -3.608 | -0.0039 | No | ||
| 86 | OSR2 | 16268 | -3.641 | 0.0061 | No | ||
| 87 | TAF5L | 16301 | -3.681 | 0.0153 | No | ||
| 88 | TMOD3 | 16540 | -4.066 | 0.0146 | No | ||
| 89 | NEDD4 | 16589 | -4.146 | 0.0244 | No | ||
| 90 | PDE7A | 16842 | -4.656 | 0.0246 | No | ||
| 91 | SEMA5B | 17229 | -5.557 | 0.0204 | No | ||
| 92 | HOXC4 | 17327 | -5.782 | 0.0324 | No | ||
| 93 | HEPH | 17388 | -5.978 | 0.0470 | No | ||
| 94 | LMO4 | 17529 | -6.463 | 0.0587 | No |