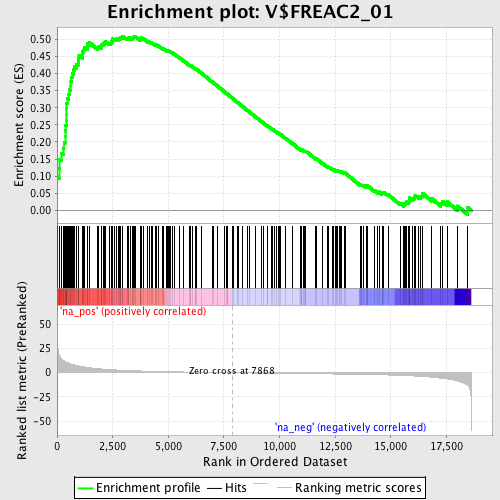
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FREAC2_01 |
| Enrichment Score (ES) | 0.5091663 |
| Normalized Enrichment Score (NES) | 1.8640695 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.040424097 |
| FWER p-Value | 0.073 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 63.431 | 0.0991 | Yes | ||
| 2 | PRKCH | 92 | 18.399 | 0.1229 | Yes | ||
| 3 | CDKN1A | 113 | 17.385 | 0.1489 | Yes | ||
| 4 | MARCKS | 184 | 14.244 | 0.1674 | Yes | ||
| 5 | NDRG1 | 274 | 12.584 | 0.1822 | Yes | ||
| 6 | SMAD1 | 309 | 12.223 | 0.1994 | Yes | ||
| 7 | SQSTM1 | 358 | 11.554 | 0.2149 | Yes | ||
| 8 | BCL11A | 362 | 11.533 | 0.2327 | Yes | ||
| 9 | SEPP1 | 380 | 11.188 | 0.2493 | Yes | ||
| 10 | HBP1 | 422 | 10.739 | 0.2638 | Yes | ||
| 11 | ERG | 426 | 10.702 | 0.2804 | Yes | ||
| 12 | KLF3 | 428 | 10.680 | 0.2970 | Yes | ||
| 13 | TIAM1 | 431 | 10.657 | 0.3135 | Yes | ||
| 14 | MBNL2 | 470 | 10.212 | 0.3274 | Yes | ||
| 15 | NFIX | 523 | 9.724 | 0.3398 | Yes | ||
| 16 | PPAP2B | 545 | 9.450 | 0.3534 | Yes | ||
| 17 | MMP13 | 619 | 8.851 | 0.3633 | Yes | ||
| 18 | ABTB1 | 622 | 8.824 | 0.3769 | Yes | ||
| 19 | THRA | 657 | 8.562 | 0.3884 | Yes | ||
| 20 | RAB6IP1 | 677 | 8.457 | 0.4006 | Yes | ||
| 21 | SMAD5 | 733 | 8.146 | 0.4104 | Yes | ||
| 22 | HOXB4 | 768 | 7.921 | 0.4209 | Yes | ||
| 23 | LRP5 | 863 | 7.384 | 0.4273 | Yes | ||
| 24 | ETV5 | 941 | 7.015 | 0.4341 | Yes | ||
| 25 | MBNL1 | 963 | 6.901 | 0.4437 | Yes | ||
| 26 | UHRF2 | 982 | 6.819 | 0.4534 | Yes | ||
| 27 | AXL | 1143 | 6.168 | 0.4544 | Yes | ||
| 28 | CHD2 | 1148 | 6.153 | 0.4638 | Yes | ||
| 29 | SH3GL3 | 1207 | 5.894 | 0.4698 | Yes | ||
| 30 | IGF1 | 1238 | 5.779 | 0.4772 | Yes | ||
| 31 | DUSP1 | 1361 | 5.322 | 0.4789 | Yes | ||
| 32 | TBCC | 1372 | 5.304 | 0.4866 | Yes | ||
| 33 | ELMO1 | 1445 | 5.102 | 0.4907 | Yes | ||
| 34 | PAK1 | 1819 | 4.193 | 0.4770 | Yes | ||
| 35 | PITPNC1 | 1880 | 4.070 | 0.4801 | Yes | ||
| 36 | EGLN1 | 1990 | 3.839 | 0.4802 | Yes | ||
| 37 | NAP1L3 | 2005 | 3.809 | 0.4854 | Yes | ||
| 38 | TCF15 | 2065 | 3.695 | 0.4880 | Yes | ||
| 39 | PELI2 | 2139 | 3.557 | 0.4896 | Yes | ||
| 40 | BRIP1 | 2165 | 3.530 | 0.4937 | Yes | ||
| 41 | FBXW7 | 2351 | 3.213 | 0.4887 | Yes | ||
| 42 | EYA1 | 2422 | 3.111 | 0.4898 | Yes | ||
| 43 | HIBADH | 2455 | 3.072 | 0.4928 | Yes | ||
| 44 | ARID4A | 2478 | 3.041 | 0.4964 | Yes | ||
| 45 | UTX | 2480 | 3.038 | 0.5011 | Yes | ||
| 46 | PPM1D | 2574 | 2.889 | 0.5005 | Yes | ||
| 47 | CALD1 | 2646 | 2.800 | 0.5011 | Yes | ||
| 48 | PLAG1 | 2740 | 2.688 | 0.5002 | Yes | ||
| 49 | DUSP4 | 2783 | 2.646 | 0.5021 | Yes | ||
| 50 | DHRS3 | 2850 | 2.563 | 0.5025 | Yes | ||
| 51 | KBTBD2 | 2858 | 2.547 | 0.5061 | Yes | ||
| 52 | RPS18 | 2931 | 2.469 | 0.5060 | Yes | ||
| 53 | IL6ST | 2945 | 2.449 | 0.5092 | Yes | ||
| 54 | HIPK1 | 3149 | 2.247 | 0.5017 | No | ||
| 55 | STOML3 | 3210 | 2.180 | 0.5018 | No | ||
| 56 | ZADH2 | 3227 | 2.164 | 0.5043 | No | ||
| 57 | PLCB2 | 3305 | 2.098 | 0.5034 | No | ||
| 58 | VPS52 | 3370 | 2.040 | 0.5031 | No | ||
| 59 | PCF11 | 3408 | 2.006 | 0.5043 | No | ||
| 60 | CDKN1C | 3423 | 1.989 | 0.5066 | No | ||
| 61 | BCL9 | 3470 | 1.955 | 0.5072 | No | ||
| 62 | HEBP2 | 3501 | 1.934 | 0.5086 | No | ||
| 63 | PITX2 | 3728 | 1.764 | 0.4991 | No | ||
| 64 | MAP4K5 | 3731 | 1.762 | 0.5017 | No | ||
| 65 | IRS4 | 3765 | 1.736 | 0.5026 | No | ||
| 66 | TCF1 | 3777 | 1.726 | 0.5047 | No | ||
| 67 | UBE2H | 3862 | 1.664 | 0.5028 | No | ||
| 68 | PHF15 | 4070 | 1.543 | 0.4939 | No | ||
| 69 | RBM16 | 4149 | 1.503 | 0.4921 | No | ||
| 70 | CPNE1 | 4234 | 1.445 | 0.4898 | No | ||
| 71 | PIK3C2A | 4295 | 1.411 | 0.4887 | No | ||
| 72 | DNAJB12 | 4399 | 1.349 | 0.4852 | No | ||
| 73 | NGEF | 4464 | 1.306 | 0.4838 | No | ||
| 74 | RORA | 4557 | 1.246 | 0.4808 | No | ||
| 75 | MITF | 4754 | 1.153 | 0.4719 | No | ||
| 76 | NRAP | 4775 | 1.143 | 0.4726 | No | ||
| 77 | BCAS3 | 4900 | 1.085 | 0.4676 | No | ||
| 78 | RASD1 | 4974 | 1.048 | 0.4653 | No | ||
| 79 | TRPC4 | 4988 | 1.043 | 0.4662 | No | ||
| 80 | NFRKB | 5035 | 1.019 | 0.4653 | No | ||
| 81 | VIT | 5114 | 0.986 | 0.4626 | No | ||
| 82 | EZH1 | 5185 | 0.954 | 0.4603 | No | ||
| 83 | GNPNAT1 | 5273 | 0.910 | 0.4570 | No | ||
| 84 | HAND2 | 5497 | 0.819 | 0.4462 | No | ||
| 85 | FGF12 | 5688 | 0.738 | 0.4370 | No | ||
| 86 | ACCN1 | 5961 | 0.642 | 0.4233 | No | ||
| 87 | AP1G2 | 5994 | 0.628 | 0.4225 | No | ||
| 88 | EXT1 | 5998 | 0.627 | 0.4233 | No | ||
| 89 | HOXA11 | 6086 | 0.585 | 0.4195 | No | ||
| 90 | KCTD15 | 6197 | 0.545 | 0.4144 | No | ||
| 91 | STK3 | 6237 | 0.529 | 0.4131 | No | ||
| 92 | PCDH18 | 6259 | 0.521 | 0.4128 | No | ||
| 93 | BRUNOL6 | 6468 | 0.445 | 0.4022 | No | ||
| 94 | SLC2A4 | 6969 | 0.278 | 0.3755 | No | ||
| 95 | ATBF1 | 7019 | 0.260 | 0.3733 | No | ||
| 96 | MRGPRF | 7217 | 0.195 | 0.3629 | No | ||
| 97 | BMP2 | 7510 | 0.111 | 0.3472 | No | ||
| 98 | MLLT6 | 7613 | 0.078 | 0.3418 | No | ||
| 99 | DHX8 | 7653 | 0.063 | 0.3398 | No | ||
| 100 | SYT1 | 7677 | 0.056 | 0.3387 | No | ||
| 101 | HHIP | 7886 | -0.005 | 0.3274 | No | ||
| 102 | ENPP2 | 7917 | -0.013 | 0.3258 | No | ||
| 103 | GPR4 | 8094 | -0.073 | 0.3163 | No | ||
| 104 | SCRN3 | 8158 | -0.090 | 0.3131 | No | ||
| 105 | PLUNC | 8326 | -0.141 | 0.3042 | No | ||
| 106 | ROR1 | 8577 | -0.214 | 0.2910 | No | ||
| 107 | CLDN8 | 8625 | -0.232 | 0.2888 | No | ||
| 108 | PURA | 8934 | -0.326 | 0.2726 | No | ||
| 109 | SIN3A | 9176 | -0.392 | 0.2602 | No | ||
| 110 | CD2AP | 9269 | -0.421 | 0.2558 | No | ||
| 111 | DSTN | 9436 | -0.468 | 0.2476 | No | ||
| 112 | HOXD9 | 9456 | -0.474 | 0.2473 | No | ||
| 113 | TGFB3 | 9645 | -0.526 | 0.2379 | No | ||
| 114 | RARA | 9678 | -0.536 | 0.2370 | No | ||
| 115 | CITED2 | 9749 | -0.557 | 0.2341 | No | ||
| 116 | PAPPA | 9881 | -0.596 | 0.2279 | No | ||
| 117 | GREM1 | 9928 | -0.609 | 0.2264 | No | ||
| 118 | RAD21 | 9989 | -0.624 | 0.2241 | No | ||
| 119 | SLC34A3 | 10051 | -0.644 | 0.2218 | No | ||
| 120 | CGA | 10285 | -0.703 | 0.2102 | No | ||
| 121 | SLITRK5 | 10589 | -0.791 | 0.1950 | No | ||
| 122 | SYT6 | 10917 | -0.882 | 0.1787 | No | ||
| 123 | FOXP2 | 10976 | -0.896 | 0.1769 | No | ||
| 124 | EN1 | 11004 | -0.905 | 0.1769 | No | ||
| 125 | SLC38A3 | 11072 | -0.922 | 0.1747 | No | ||
| 126 | WEE1 | 11125 | -0.938 | 0.1733 | No | ||
| 127 | EVI1 | 11169 | -0.951 | 0.1725 | No | ||
| 128 | RASGEF1B | 11178 | -0.952 | 0.1735 | No | ||
| 129 | FGF9 | 11632 | -1.082 | 0.1507 | No | ||
| 130 | HABP4 | 11673 | -1.098 | 0.1502 | No | ||
| 131 | MID1 | 11932 | -1.176 | 0.1380 | No | ||
| 132 | PCSK1 | 12166 | -1.250 | 0.1274 | No | ||
| 133 | MCC | 12218 | -1.264 | 0.1266 | No | ||
| 134 | BMP5 | 12366 | -1.309 | 0.1206 | No | ||
| 135 | EPHB3 | 12426 | -1.327 | 0.1195 | No | ||
| 136 | KLF12 | 12519 | -1.353 | 0.1166 | No | ||
| 137 | BCL11B | 12551 | -1.361 | 0.1171 | No | ||
| 138 | CD109 | 12587 | -1.370 | 0.1173 | No | ||
| 139 | POU3F3 | 12677 | -1.403 | 0.1147 | No | ||
| 140 | NRG1 | 12758 | -1.429 | 0.1126 | No | ||
| 141 | NFIB | 12804 | -1.445 | 0.1124 | No | ||
| 142 | BDNF | 12898 | -1.477 | 0.1097 | No | ||
| 143 | FGF20 | 12944 | -1.491 | 0.1095 | No | ||
| 144 | EN2 | 13623 | -1.738 | 0.0755 | No | ||
| 145 | ADAM12 | 13700 | -1.770 | 0.0741 | No | ||
| 146 | AP4M1 | 13761 | -1.795 | 0.0737 | No | ||
| 147 | HOXC12 | 13890 | -1.848 | 0.0696 | No | ||
| 148 | FOXB1 | 13933 | -1.869 | 0.0703 | No | ||
| 149 | CHCHD7 | 13938 | -1.871 | 0.0730 | No | ||
| 150 | OFCC1 | 14269 | -2.020 | 0.0582 | No | ||
| 151 | CXXC5 | 14409 | -2.096 | 0.0540 | No | ||
| 152 | HR | 14469 | -2.130 | 0.0541 | No | ||
| 153 | RBM4 | 14603 | -2.193 | 0.0503 | No | ||
| 154 | MAFF | 14629 | -2.209 | 0.0524 | No | ||
| 155 | E2F3 | 14675 | -2.239 | 0.0534 | No | ||
| 156 | TBX4 | 14874 | -2.350 | 0.0464 | No | ||
| 157 | DSG4 | 15422 | -2.735 | 0.0210 | No | ||
| 158 | CLPX | 15589 | -2.866 | 0.0165 | No | ||
| 159 | TPP2 | 15625 | -2.897 | 0.0191 | No | ||
| 160 | DTNA | 15663 | -2.927 | 0.0216 | No | ||
| 161 | PRDM1 | 15689 | -2.948 | 0.0249 | No | ||
| 162 | TOB1 | 15802 | -3.090 | 0.0236 | No | ||
| 163 | GPRC5C | 15830 | -3.125 | 0.0271 | No | ||
| 164 | PROX1 | 15833 | -3.126 | 0.0318 | No | ||
| 165 | CDX2 | 15835 | -3.128 | 0.0367 | No | ||
| 166 | NNT | 15954 | -3.247 | 0.0353 | No | ||
| 167 | FOSL2 | 16044 | -3.351 | 0.0357 | No | ||
| 168 | AKT2 | 16062 | -3.372 | 0.0401 | No | ||
| 169 | HOXC8 | 16107 | -3.422 | 0.0430 | No | ||
| 170 | STOML2 | 16245 | -3.603 | 0.0412 | No | ||
| 171 | POU3F4 | 16353 | -3.758 | 0.0413 | No | ||
| 172 | MCM7 | 16403 | -3.842 | 0.0446 | No | ||
| 173 | MRVI1 | 16434 | -3.889 | 0.0491 | No | ||
| 174 | PDE7A | 16842 | -4.656 | 0.0343 | No | ||
| 175 | MPP6 | 17249 | -5.598 | 0.0210 | No | ||
| 176 | HOXC4 | 17327 | -5.782 | 0.0259 | No | ||
| 177 | LMO4 | 17529 | -6.463 | 0.0251 | No | ||
| 178 | DOCK3 | 17998 | -8.531 | 0.0130 | No | ||
| 179 | CPN1 | 18457 | -13.108 | 0.0086 | No |