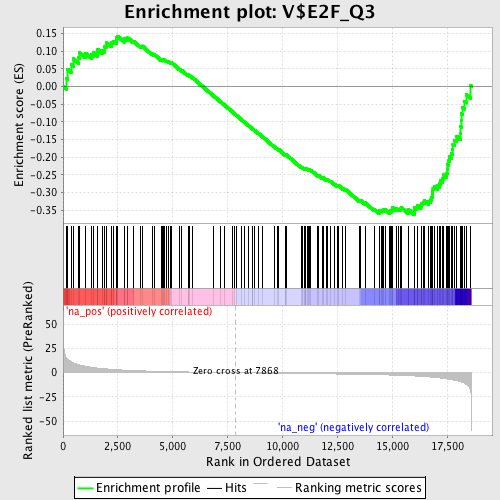
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$E2F_Q3 |
| Enrichment Score (ES) | -0.36180195 |
| Normalized Enrichment Score (NES) | -1.3208308 |
| Nominal p-value | 0.036170214 |
| FDR q-value | 0.3752082 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MXD3 | 141 | 15.681 | 0.0238 | No | ||
| 2 | LASP1 | 206 | 13.841 | 0.0480 | No | ||
| 3 | DCK | 364 | 11.502 | 0.0626 | No | ||
| 4 | MEIS1 | 458 | 10.375 | 0.0783 | No | ||
| 5 | RAB11B | 716 | 8.273 | 0.0810 | No | ||
| 6 | CTCF | 758 | 8.038 | 0.0949 | No | ||
| 7 | DMD | 1013 | 6.671 | 0.0945 | No | ||
| 8 | SSBP3 | 1302 | 5.517 | 0.0899 | No | ||
| 9 | ZCCHC8 | 1398 | 5.228 | 0.0952 | No | ||
| 10 | EZH2 | 1576 | 4.762 | 0.0952 | No | ||
| 11 | DPYSL2 | 1581 | 4.741 | 0.1045 | No | ||
| 12 | PIAS1 | 1786 | 4.255 | 0.1020 | No | ||
| 13 | TBX6 | 1876 | 4.078 | 0.1053 | No | ||
| 14 | PHF12 | 1877 | 4.077 | 0.1135 | No | ||
| 15 | PIM1 | 1960 | 3.901 | 0.1168 | No | ||
| 16 | HIST1H2BK | 1981 | 3.860 | 0.1235 | No | ||
| 17 | PAPOLG | 2197 | 3.489 | 0.1188 | No | ||
| 18 | EPHB2 | 2209 | 3.475 | 0.1252 | No | ||
| 19 | CTDSP1 | 2287 | 3.323 | 0.1277 | No | ||
| 20 | DDB2 | 2429 | 3.104 | 0.1263 | No | ||
| 21 | E2F7 | 2444 | 3.084 | 0.1317 | No | ||
| 22 | KLF4 | 2449 | 3.079 | 0.1376 | No | ||
| 23 | ARID4A | 2478 | 3.041 | 0.1422 | No | ||
| 24 | ARHGAP6 | 2790 | 2.639 | 0.1307 | No | ||
| 25 | KLF5 | 2803 | 2.624 | 0.1353 | No | ||
| 26 | PRKDC | 2930 | 2.469 | 0.1334 | No | ||
| 27 | ERBB2IP | 2952 | 2.443 | 0.1372 | No | ||
| 28 | GCH1 | 3201 | 2.189 | 0.1281 | No | ||
| 29 | AP1S2 | 3546 | 1.902 | 0.1133 | No | ||
| 30 | OLFML3 | 3597 | 1.862 | 0.1143 | No | ||
| 31 | PHF15 | 4070 | 1.543 | 0.0919 | No | ||
| 32 | DNMT1 | 4168 | 1.490 | 0.0896 | No | ||
| 33 | APRIN | 4476 | 1.301 | 0.0756 | No | ||
| 34 | CBX5 | 4545 | 1.254 | 0.0744 | No | ||
| 35 | NUP153 | 4572 | 1.239 | 0.0755 | No | ||
| 36 | ADAMTS2 | 4629 | 1.208 | 0.0749 | No | ||
| 37 | HIST1H2AH | 4730 | 1.167 | 0.0718 | No | ||
| 38 | SALL1 | 4814 | 1.124 | 0.0695 | No | ||
| 39 | H2AFZ | 4915 | 1.079 | 0.0663 | No | ||
| 40 | ATRX | 4929 | 1.072 | 0.0677 | No | ||
| 41 | HMGN2 | 5302 | 0.896 | 0.0494 | No | ||
| 42 | MAZ | 5416 | 0.852 | 0.0450 | No | ||
| 43 | FMO4 | 5698 | 0.732 | 0.0312 | No | ||
| 44 | FLI1 | 5702 | 0.729 | 0.0325 | No | ||
| 45 | USP1 | 5770 | 0.708 | 0.0303 | No | ||
| 46 | STK35 | 5900 | 0.666 | 0.0247 | No | ||
| 47 | RHD | 6870 | 0.307 | -0.0272 | No | ||
| 48 | FBXO5 | 7171 | 0.211 | -0.0430 | No | ||
| 49 | HNRPD | 7344 | 0.156 | -0.0520 | No | ||
| 50 | CASP8AP2 | 7699 | 0.048 | -0.0711 | No | ||
| 51 | POLE2 | 7824 | 0.012 | -0.0778 | No | ||
| 52 | CPNE5 | 7907 | -0.011 | -0.0822 | No | ||
| 53 | PEG3 | 8144 | -0.087 | -0.0948 | No | ||
| 54 | RFC1 | 8267 | -0.124 | -0.1011 | No | ||
| 55 | SLC38A1 | 8443 | -0.174 | -0.1103 | No | ||
| 56 | ADCY8 | 8632 | -0.234 | -0.1200 | No | ||
| 57 | BARHL1 | 8636 | -0.235 | -0.1197 | No | ||
| 58 | ASCL1 | 8723 | -0.263 | -0.1238 | No | ||
| 59 | HS6ST3 | 8910 | -0.317 | -0.1332 | No | ||
| 60 | SFRS1 | 9103 | -0.371 | -0.1429 | No | ||
| 61 | STAG2 | 9636 | -0.524 | -0.1706 | No | ||
| 62 | HIST1H1D | 9770 | -0.563 | -0.1767 | No | ||
| 63 | NPR3 | 9830 | -0.582 | -0.1787 | No | ||
| 64 | TOPBP1 | 10139 | -0.666 | -0.1941 | No | ||
| 65 | PCNA | 10147 | -0.668 | -0.1931 | No | ||
| 66 | BAT3 | 10175 | -0.675 | -0.1932 | No | ||
| 67 | EPHB1 | 10854 | -0.863 | -0.2282 | No | ||
| 68 | CLTA | 10912 | -0.881 | -0.2295 | No | ||
| 69 | TREX2 | 10992 | -0.900 | -0.2320 | No | ||
| 70 | LHX5 | 11035 | -0.913 | -0.2325 | No | ||
| 71 | SLC25A3 | 11062 | -0.919 | -0.2320 | No | ||
| 72 | WEE1 | 11125 | -0.938 | -0.2335 | No | ||
| 73 | TRIM47 | 11193 | -0.955 | -0.2352 | No | ||
| 74 | PLK4 | 11212 | -0.962 | -0.2343 | No | ||
| 75 | MDGA1 | 11291 | -0.985 | -0.2365 | No | ||
| 76 | STMN1 | 11601 | -1.074 | -0.2511 | No | ||
| 77 | SLC6A4 | 11656 | -1.093 | -0.2518 | No | ||
| 78 | SEZ6 | 11820 | -1.143 | -0.2584 | No | ||
| 79 | IPO7 | 11849 | -1.155 | -0.2576 | No | ||
| 80 | TBX3 | 12008 | -1.199 | -0.2637 | No | ||
| 81 | HNRPA1 | 12031 | -1.207 | -0.2625 | No | ||
| 82 | PCSK1 | 12166 | -1.250 | -0.2672 | No | ||
| 83 | NOL4 | 12375 | -1.311 | -0.2759 | No | ||
| 84 | PHF13 | 12505 | -1.351 | -0.2802 | No | ||
| 85 | SNPH | 12538 | -1.357 | -0.2792 | No | ||
| 86 | MCM8 | 12751 | -1.427 | -0.2878 | No | ||
| 87 | PODN | 12879 | -1.471 | -0.2917 | No | ||
| 88 | NDUFA11 | 13513 | -1.697 | -0.3226 | No | ||
| 89 | ATAD2 | 13560 | -1.718 | -0.3216 | No | ||
| 90 | AP4M1 | 13761 | -1.795 | -0.3289 | No | ||
| 91 | LIG1 | 14187 | -1.982 | -0.3479 | No | ||
| 92 | NRP2 | 14401 | -2.091 | -0.3553 | Yes | ||
| 93 | ONECUT1 | 14410 | -2.096 | -0.3515 | Yes | ||
| 94 | RBBP4 | 14492 | -2.139 | -0.3516 | Yes | ||
| 95 | CACNA1G | 14540 | -2.167 | -0.3498 | Yes | ||
| 96 | GRIK2 | 14594 | -2.191 | -0.3483 | Yes | ||
| 97 | E2F3 | 14675 | -2.239 | -0.3481 | Yes | ||
| 98 | UXT | 14887 | -2.355 | -0.3548 | Yes | ||
| 99 | PAQR4 | 14914 | -2.374 | -0.3515 | Yes | ||
| 100 | TFRC | 14981 | -2.412 | -0.3502 | Yes | ||
| 101 | DLST | 15014 | -2.439 | -0.3471 | Yes | ||
| 102 | CNOT3 | 15023 | -2.443 | -0.3426 | Yes | ||
| 103 | CDCA7 | 15172 | -2.549 | -0.3455 | Yes | ||
| 104 | NCL | 15287 | -2.635 | -0.3464 | Yes | ||
| 105 | MCM2 | 15360 | -2.690 | -0.3449 | Yes | ||
| 106 | SYT11 | 15420 | -2.735 | -0.3426 | Yes | ||
| 107 | MCM6 | 15736 | -3.002 | -0.3537 | Yes | ||
| 108 | INSM1 | 15737 | -3.003 | -0.3477 | Yes | ||
| 109 | SEMA5A | 15999 | -3.291 | -0.3552 | Yes | ||
| 110 | UFD1L | 16010 | -3.308 | -0.3491 | Yes | ||
| 111 | JPH1 | 16014 | -3.313 | -0.3427 | Yes | ||
| 112 | SLC25A14 | 16130 | -3.443 | -0.3420 | Yes | ||
| 113 | PRPS1 | 16142 | -3.454 | -0.3357 | Yes | ||
| 114 | MCM4 | 16312 | -3.695 | -0.3374 | Yes | ||
| 115 | E2F1 | 16330 | -3.733 | -0.3309 | Yes | ||
| 116 | MCM7 | 16403 | -3.842 | -0.3271 | Yes | ||
| 117 | CDC25A | 16478 | -3.968 | -0.3231 | Yes | ||
| 118 | KCNA1 | 16665 | -4.283 | -0.3246 | Yes | ||
| 119 | PVRL1 | 16750 | -4.458 | -0.3202 | Yes | ||
| 120 | MCM3 | 16797 | -4.561 | -0.3136 | Yes | ||
| 121 | PCSK4 | 16821 | -4.617 | -0.3056 | Yes | ||
| 122 | ATE1 | 16837 | -4.644 | -0.2971 | Yes | ||
| 123 | GMNN | 16856 | -4.684 | -0.2887 | Yes | ||
| 124 | NASP | 16906 | -4.765 | -0.2818 | Yes | ||
| 125 | KCND2 | 17078 | -5.144 | -0.2807 | Yes | ||
| 126 | DNAJC11 | 17169 | -5.388 | -0.2748 | Yes | ||
| 127 | POLA2 | 17214 | -5.514 | -0.2662 | Yes | ||
| 128 | SMAD6 | 17306 | -5.728 | -0.2596 | Yes | ||
| 129 | RBBP7 | 17347 | -5.853 | -0.2501 | Yes | ||
| 130 | PPP1R8 | 17490 | -6.340 | -0.2450 | Yes | ||
| 131 | MAT2A | 17508 | -6.409 | -0.2331 | Yes | ||
| 132 | KPNB1 | 17532 | -6.478 | -0.2214 | Yes | ||
| 133 | PPP1CC | 17571 | -6.598 | -0.2102 | Yes | ||
| 134 | CDC6 | 17598 | -6.709 | -0.1982 | Yes | ||
| 135 | HTF9C | 17693 | -7.058 | -0.1892 | Yes | ||
| 136 | SFRS2 | 17755 | -7.315 | -0.1778 | Yes | ||
| 137 | CDC45L | 17767 | -7.357 | -0.1637 | Yes | ||
| 138 | ING3 | 17844 | -7.748 | -0.1523 | Yes | ||
| 139 | ORC1L | 17922 | -8.076 | -0.1403 | Yes | ||
| 140 | UNG | 18103 | -9.207 | -0.1316 | Yes | ||
| 141 | RAD51 | 18119 | -9.329 | -0.1137 | Yes | ||
| 142 | RANBP1 | 18156 | -9.677 | -0.0963 | Yes | ||
| 143 | TCP1 | 18164 | -9.742 | -0.0771 | Yes | ||
| 144 | MRPL18 | 18195 | -9.981 | -0.0588 | Yes | ||
| 145 | IMPDH2 | 18290 | -10.804 | -0.0422 | Yes | ||
| 146 | ABCF2 | 18376 | -11.997 | -0.0228 | Yes | ||
| 147 | SHMT1 | 18568 | -17.838 | 0.0026 | Yes |
