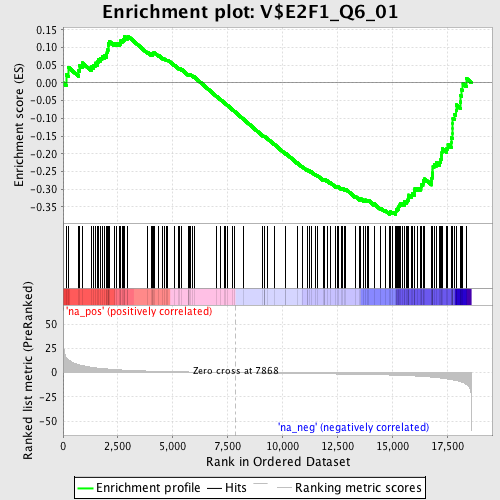
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$E2F1_Q6_01 |
| Enrichment Score (ES) | -0.37131277 |
| Normalized Enrichment Score (NES) | -1.3721932 |
| Nominal p-value | 0.010893246 |
| FDR q-value | 0.29147476 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MXD3 | 141 | 15.681 | 0.0232 | No | ||
| 2 | GPRC5B | 242 | 13.296 | 0.0440 | No | ||
| 3 | RAB11B | 716 | 8.273 | 0.0346 | No | ||
| 4 | CTCF | 758 | 8.038 | 0.0482 | No | ||
| 5 | EFNA5 | 872 | 7.329 | 0.0565 | No | ||
| 6 | CASP2 | 1282 | 5.602 | 0.0454 | No | ||
| 7 | FHOD1 | 1386 | 5.267 | 0.0502 | No | ||
| 8 | PIK3CD | 1460 | 5.066 | 0.0562 | No | ||
| 9 | EZH2 | 1576 | 4.762 | 0.0593 | No | ||
| 10 | HNRPUL1 | 1600 | 4.688 | 0.0673 | No | ||
| 11 | POU2F1 | 1708 | 4.424 | 0.0702 | No | ||
| 12 | CCNT1 | 1773 | 4.279 | 0.0752 | No | ||
| 13 | TBX6 | 1876 | 4.078 | 0.0777 | No | ||
| 14 | HIST1H2BK | 1981 | 3.860 | 0.0796 | No | ||
| 15 | POLD3 | 1999 | 3.822 | 0.0862 | No | ||
| 16 | USP2 | 2010 | 3.801 | 0.0932 | No | ||
| 17 | CHD4 | 2049 | 3.732 | 0.0985 | No | ||
| 18 | SLC25A1 | 2050 | 3.728 | 0.1058 | No | ||
| 19 | NUMA1 | 2062 | 3.697 | 0.1125 | No | ||
| 20 | RBL1 | 2116 | 3.612 | 0.1167 | No | ||
| 21 | NCOA6 | 2343 | 3.229 | 0.1108 | No | ||
| 22 | DDB2 | 2429 | 3.104 | 0.1123 | No | ||
| 23 | PPM1D | 2574 | 2.889 | 0.1102 | No | ||
| 24 | ILF3 | 2615 | 2.843 | 0.1137 | No | ||
| 25 | USP52 | 2629 | 2.819 | 0.1185 | No | ||
| 26 | TRIM39 | 2692 | 2.744 | 0.1205 | No | ||
| 27 | SPHK2 | 2746 | 2.686 | 0.1230 | No | ||
| 28 | ARHGAP6 | 2790 | 2.639 | 0.1258 | No | ||
| 29 | KLF5 | 2803 | 2.624 | 0.1303 | No | ||
| 30 | PRKDC | 2930 | 2.469 | 0.1284 | No | ||
| 31 | ERBB2IP | 2952 | 2.443 | 0.1320 | No | ||
| 32 | TNF | 3842 | 1.682 | 0.0872 | No | ||
| 33 | STAG1 | 4041 | 1.558 | 0.0795 | No | ||
| 34 | PHF15 | 4070 | 1.543 | 0.0810 | No | ||
| 35 | PTMA | 4071 | 1.542 | 0.0841 | No | ||
| 36 | MYH10 | 4116 | 1.519 | 0.0847 | No | ||
| 37 | DNMT1 | 4168 | 1.490 | 0.0849 | No | ||
| 38 | BRMS1L | 4351 | 1.379 | 0.0777 | No | ||
| 39 | CBX5 | 4545 | 1.254 | 0.0697 | No | ||
| 40 | ADAMTS2 | 4629 | 1.208 | 0.0676 | No | ||
| 41 | HIST1H2AH | 4730 | 1.167 | 0.0645 | No | ||
| 42 | SP1 | 4770 | 1.146 | 0.0646 | No | ||
| 43 | DHH | 5073 | 1.004 | 0.0502 | No | ||
| 44 | DNAJC9 | 5280 | 0.907 | 0.0409 | No | ||
| 45 | HMGN2 | 5302 | 0.896 | 0.0415 | No | ||
| 46 | MAZ | 5416 | 0.852 | 0.0370 | No | ||
| 47 | FLI1 | 5702 | 0.729 | 0.0230 | No | ||
| 48 | GNG4 | 5706 | 0.727 | 0.0243 | No | ||
| 49 | NPAT | 5720 | 0.723 | 0.0250 | No | ||
| 50 | MGAT2 | 5773 | 0.707 | 0.0236 | No | ||
| 51 | TNFRSF19L | 5789 | 0.704 | 0.0242 | No | ||
| 52 | STK35 | 5900 | 0.666 | 0.0195 | No | ||
| 53 | FZD1 | 5965 | 0.639 | 0.0173 | No | ||
| 54 | TMPO | 6996 | 0.267 | -0.0380 | No | ||
| 55 | FBXO5 | 7171 | 0.211 | -0.0470 | No | ||
| 56 | HNRPD | 7344 | 0.156 | -0.0560 | No | ||
| 57 | FKBP5 | 7406 | 0.139 | -0.0590 | No | ||
| 58 | BMP2 | 7510 | 0.111 | -0.0644 | No | ||
| 59 | CASP8AP2 | 7699 | 0.048 | -0.0745 | No | ||
| 60 | POLE2 | 7824 | 0.012 | -0.0812 | No | ||
| 61 | FANCG | 8202 | -0.102 | -0.1014 | No | ||
| 62 | SFRS1 | 9103 | -0.371 | -0.1494 | No | ||
| 63 | REPS2 | 9157 | -0.386 | -0.1515 | No | ||
| 64 | SIN3A | 9176 | -0.392 | -0.1517 | No | ||
| 65 | KCNS2 | 9180 | -0.393 | -0.1511 | No | ||
| 66 | ASXL2 | 9300 | -0.428 | -0.1567 | No | ||
| 67 | STAG2 | 9636 | -0.524 | -0.1738 | No | ||
| 68 | TOPBP1 | 10139 | -0.666 | -0.1997 | No | ||
| 69 | PCNA | 10147 | -0.668 | -0.1988 | No | ||
| 70 | RPL18 | 10664 | -0.814 | -0.2252 | No | ||
| 71 | SYT6 | 10917 | -0.882 | -0.2371 | No | ||
| 72 | WEE1 | 11125 | -0.938 | -0.2464 | No | ||
| 73 | PAX6 | 11145 | -0.944 | -0.2456 | No | ||
| 74 | SMARCA1 | 11220 | -0.964 | -0.2477 | No | ||
| 75 | TYRO3 | 11299 | -0.987 | -0.2500 | No | ||
| 76 | HCN3 | 11503 | -1.044 | -0.2589 | No | ||
| 77 | STMN1 | 11601 | -1.074 | -0.2621 | No | ||
| 78 | IPO7 | 11849 | -1.155 | -0.2732 | No | ||
| 79 | RB1CC1 | 11909 | -1.169 | -0.2741 | No | ||
| 80 | ERF | 11926 | -1.175 | -0.2726 | No | ||
| 81 | HNRPA1 | 12031 | -1.207 | -0.2759 | No | ||
| 82 | PCSK1 | 12166 | -1.250 | -0.2807 | No | ||
| 83 | ENPP1 | 12433 | -1.328 | -0.2925 | No | ||
| 84 | PHF13 | 12505 | -1.351 | -0.2937 | No | ||
| 85 | CLSPN | 12529 | -1.355 | -0.2923 | No | ||
| 86 | HOXC10 | 12681 | -1.404 | -0.2977 | No | ||
| 87 | MCM8 | 12751 | -1.427 | -0.2986 | No | ||
| 88 | SYNGR4 | 12845 | -1.459 | -0.3008 | No | ||
| 89 | PODN | 12879 | -1.471 | -0.2997 | No | ||
| 90 | IRX3 | 13322 | -1.623 | -0.3204 | No | ||
| 91 | ATP5G2 | 13518 | -1.699 | -0.3277 | No | ||
| 92 | ATAD2 | 13560 | -1.718 | -0.3265 | No | ||
| 93 | ACTN3 | 13703 | -1.771 | -0.3307 | No | ||
| 94 | AP4M1 | 13761 | -1.795 | -0.3303 | No | ||
| 95 | HMGA2 | 13856 | -1.835 | -0.3317 | No | ||
| 96 | CDC5L | 13909 | -1.857 | -0.3309 | No | ||
| 97 | LRP1 | 14171 | -1.976 | -0.3412 | No | ||
| 98 | HR | 14469 | -2.130 | -0.3530 | No | ||
| 99 | E2F3 | 14675 | -2.239 | -0.3597 | No | ||
| 100 | UXT | 14887 | -2.355 | -0.3665 | Yes | ||
| 101 | PAQR4 | 14914 | -2.374 | -0.3633 | Yes | ||
| 102 | CNOT3 | 15023 | -2.443 | -0.3643 | Yes | ||
| 103 | GADD45B | 15153 | -2.533 | -0.3663 | Yes | ||
| 104 | CDCA7 | 15172 | -2.549 | -0.3623 | Yes | ||
| 105 | YBX2 | 15178 | -2.555 | -0.3575 | Yes | ||
| 106 | PCDH10 | 15237 | -2.593 | -0.3556 | Yes | ||
| 107 | NCL | 15287 | -2.635 | -0.3530 | Yes | ||
| 108 | MTSS1 | 15304 | -2.646 | -0.3487 | Yes | ||
| 109 | GATA1 | 15324 | -2.661 | -0.3445 | Yes | ||
| 110 | MCM2 | 15360 | -2.690 | -0.3411 | Yes | ||
| 111 | ACBD6 | 15466 | -2.764 | -0.3413 | Yes | ||
| 112 | MYC | 15548 | -2.832 | -0.3402 | Yes | ||
| 113 | AP1S1 | 15549 | -2.832 | -0.3346 | Yes | ||
| 114 | ACO2 | 15666 | -2.929 | -0.3351 | Yes | ||
| 115 | FANCC | 15686 | -2.947 | -0.3303 | Yes | ||
| 116 | MCM6 | 15736 | -3.002 | -0.3271 | Yes | ||
| 117 | INSM1 | 15737 | -3.003 | -0.3212 | Yes | ||
| 118 | HNRPR | 15758 | -3.024 | -0.3163 | Yes | ||
| 119 | FAF1 | 15891 | -3.176 | -0.3172 | Yes | ||
| 120 | AMD1 | 15909 | -3.197 | -0.3118 | Yes | ||
| 121 | UFD1L | 16010 | -3.308 | -0.3107 | Yes | ||
| 122 | JPH1 | 16014 | -3.313 | -0.3044 | Yes | ||
| 123 | ATM | 16035 | -3.341 | -0.2989 | Yes | ||
| 124 | PRPS1 | 16142 | -3.454 | -0.2978 | Yes | ||
| 125 | EED | 16273 | -3.647 | -0.2977 | Yes | ||
| 126 | MCM4 | 16312 | -3.695 | -0.2925 | Yes | ||
| 127 | E2F1 | 16330 | -3.733 | -0.2861 | Yes | ||
| 128 | MCM7 | 16403 | -3.842 | -0.2824 | Yes | ||
| 129 | MSH2 | 16428 | -3.879 | -0.2761 | Yes | ||
| 130 | CDC25A | 16478 | -3.968 | -0.2709 | Yes | ||
| 131 | MCM3 | 16797 | -4.561 | -0.2792 | Yes | ||
| 132 | SUMO1 | 16805 | -4.573 | -0.2705 | Yes | ||
| 133 | RPS19 | 16817 | -4.600 | -0.2621 | Yes | ||
| 134 | ATE1 | 16837 | -4.644 | -0.2540 | Yes | ||
| 135 | GMNN | 16856 | -4.684 | -0.2457 | Yes | ||
| 136 | SYNCRIP | 16857 | -4.692 | -0.2365 | Yes | ||
| 137 | NASP | 16906 | -4.765 | -0.2297 | Yes | ||
| 138 | RQCD1 | 17000 | -4.938 | -0.2250 | Yes | ||
| 139 | DNAJC11 | 17169 | -5.388 | -0.2235 | Yes | ||
| 140 | POLA2 | 17214 | -5.514 | -0.2151 | Yes | ||
| 141 | GSPT1 | 17244 | -5.577 | -0.2057 | Yes | ||
| 142 | USP37 | 17264 | -5.627 | -0.1956 | Yes | ||
| 143 | SLC9A7 | 17272 | -5.650 | -0.1849 | Yes | ||
| 144 | PPP1R8 | 17490 | -6.340 | -0.1842 | Yes | ||
| 145 | HIRA | 17541 | -6.497 | -0.1741 | Yes | ||
| 146 | HTF9C | 17693 | -7.058 | -0.1684 | Yes | ||
| 147 | SUV39H1 | 17700 | -7.078 | -0.1548 | Yes | ||
| 148 | SFRS2 | 17755 | -7.315 | -0.1433 | Yes | ||
| 149 | NOLC1 | 17759 | -7.336 | -0.1290 | Yes | ||
| 150 | PHF5A | 17762 | -7.350 | -0.1147 | Yes | ||
| 151 | CDC45L | 17767 | -7.357 | -0.1004 | Yes | ||
| 152 | PPRC1 | 17835 | -7.677 | -0.0889 | Yes | ||
| 153 | MRPL40 | 17906 | -7.977 | -0.0770 | Yes | ||
| 154 | ORC1L | 17922 | -8.076 | -0.0619 | Yes | ||
| 155 | UNG | 18103 | -9.207 | -0.0536 | Yes | ||
| 156 | RAD51 | 18119 | -9.329 | -0.0360 | Yes | ||
| 157 | RANBP1 | 18156 | -9.677 | -0.0189 | Yes | ||
| 158 | MAP4K1 | 18224 | -10.205 | -0.0025 | Yes | ||
| 159 | AK2 | 18378 | -12.015 | 0.0129 | Yes |
