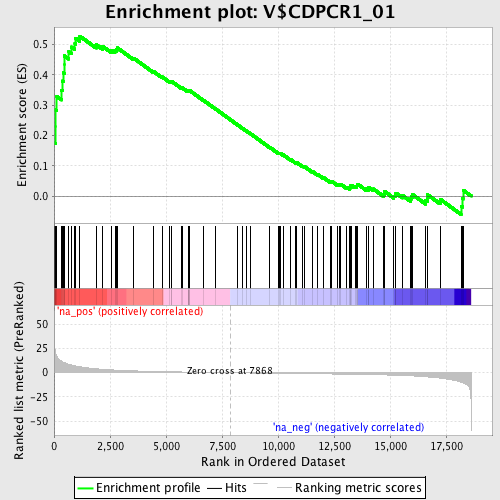
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CDPCR1_01 |
| Enrichment Score (ES) | 0.5280977 |
| Normalized Enrichment Score (NES) | 1.7864602 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.053461917 |
| FWER p-Value | 0.184 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 63.431 | 0.1765 | Yes | ||
| 2 | TCF7L2 | 72 | 20.268 | 0.2290 | Yes | ||
| 3 | IL15 | 74 | 19.919 | 0.2844 | Yes | ||
| 4 | GRB7 | 120 | 16.803 | 0.3287 | Yes | ||
| 5 | TLE4 | 336 | 11.842 | 0.3501 | Yes | ||
| 6 | BTBD3 | 370 | 11.391 | 0.3800 | Yes | ||
| 7 | ERG | 426 | 10.702 | 0.4068 | Yes | ||
| 8 | PPT2 | 455 | 10.408 | 0.4342 | Yes | ||
| 9 | MEIS1 | 458 | 10.375 | 0.4630 | Yes | ||
| 10 | THRA | 657 | 8.562 | 0.4761 | Yes | ||
| 11 | HOXB4 | 768 | 7.921 | 0.4922 | Yes | ||
| 12 | EGR1 | 929 | 7.104 | 0.5034 | Yes | ||
| 13 | MBNL1 | 963 | 6.901 | 0.5208 | Yes | ||
| 14 | FCHSD1 | 1146 | 6.160 | 0.5281 | Yes | ||
| 15 | TBX6 | 1876 | 4.078 | 0.5001 | No | ||
| 16 | ETV1 | 2167 | 3.528 | 0.4943 | No | ||
| 17 | LYPLA3 | 2573 | 2.894 | 0.4805 | No | ||
| 18 | CREM | 2719 | 2.708 | 0.4802 | No | ||
| 19 | DUSP4 | 2783 | 2.646 | 0.4841 | No | ||
| 20 | DIO2 | 2821 | 2.606 | 0.4894 | No | ||
| 21 | SERPING1 | 3564 | 1.893 | 0.4546 | No | ||
| 22 | COL2A1 | 4450 | 1.316 | 0.4105 | No | ||
| 23 | DGKB | 4827 | 1.117 | 0.3933 | No | ||
| 24 | MYST2 | 5151 | 0.970 | 0.3786 | No | ||
| 25 | NR3C2 | 5234 | 0.926 | 0.3767 | No | ||
| 26 | SON | 5246 | 0.921 | 0.3787 | No | ||
| 27 | FGF12 | 5688 | 0.738 | 0.3570 | No | ||
| 28 | PLXNA3 | 5717 | 0.725 | 0.3575 | No | ||
| 29 | NOG | 5976 | 0.637 | 0.3453 | No | ||
| 30 | EXT1 | 5998 | 0.627 | 0.3459 | No | ||
| 31 | LIN28 | 6013 | 0.622 | 0.3469 | No | ||
| 32 | PAX8 | 6047 | 0.604 | 0.3468 | No | ||
| 33 | AP3M2 | 6060 | 0.598 | 0.3478 | No | ||
| 34 | HOXC5 | 6664 | 0.378 | 0.3163 | No | ||
| 35 | NKX2-2 | 7191 | 0.205 | 0.2885 | No | ||
| 36 | PDCD10 | 8204 | -0.102 | 0.2342 | No | ||
| 37 | HOXD10 | 8430 | -0.170 | 0.2225 | No | ||
| 38 | STAT5B | 8605 | -0.224 | 0.2137 | No | ||
| 39 | ADAM10 | 8764 | -0.276 | 0.2060 | No | ||
| 40 | STAG2 | 9636 | -0.524 | 0.1604 | No | ||
| 41 | MAOB | 10013 | -0.633 | 0.1419 | No | ||
| 42 | STMN2 | 10067 | -0.648 | 0.1409 | No | ||
| 43 | VAX1 | 10097 | -0.659 | 0.1411 | No | ||
| 44 | CYP26B1 | 10225 | -0.690 | 0.1362 | No | ||
| 45 | ITGA7 | 10557 | -0.783 | 0.1205 | No | ||
| 46 | RPH3A | 10782 | -0.846 | 0.1108 | No | ||
| 47 | NRXN3 | 10804 | -0.850 | 0.1120 | No | ||
| 48 | PIGN | 11095 | -0.931 | 0.0989 | No | ||
| 49 | PFN2 | 11189 | -0.954 | 0.0966 | No | ||
| 50 | CSMD3 | 11540 | -1.057 | 0.0806 | No | ||
| 51 | NHLH2 | 11774 | -1.129 | 0.0712 | No | ||
| 52 | TBX3 | 12008 | -1.199 | 0.0620 | No | ||
| 53 | HOXC6 | 12350 | -1.302 | 0.0472 | No | ||
| 54 | NOL4 | 12375 | -1.311 | 0.0495 | No | ||
| 55 | INSRR | 12650 | -1.395 | 0.0386 | No | ||
| 56 | MGAT5B | 12748 | -1.426 | 0.0374 | No | ||
| 57 | NFIB | 12804 | -1.445 | 0.0384 | No | ||
| 58 | DPP10 | 13049 | -1.522 | 0.0295 | No | ||
| 59 | DBC1 | 13204 | -1.577 | 0.0256 | No | ||
| 60 | SLC12A5 | 13211 | -1.579 | 0.0296 | No | ||
| 61 | KCND1 | 13220 | -1.582 | 0.0336 | No | ||
| 62 | NTNG2 | 13274 | -1.602 | 0.0352 | No | ||
| 63 | GRIK4 | 13433 | -1.667 | 0.0313 | No | ||
| 64 | BHLHB3 | 13491 | -1.689 | 0.0329 | No | ||
| 65 | NTRK1 | 13521 | -1.700 | 0.0361 | No | ||
| 66 | KLF15 | 13553 | -1.715 | 0.0392 | No | ||
| 67 | RFX4 | 13931 | -1.867 | 0.0240 | No | ||
| 68 | INSM2 | 14022 | -1.910 | 0.0245 | No | ||
| 69 | CPNE6 | 14045 | -1.917 | 0.0286 | No | ||
| 70 | ST7 | 14246 | -2.013 | 0.0234 | No | ||
| 71 | HOXA10 | 14722 | -2.262 | 0.0041 | No | ||
| 72 | CUL2 | 14757 | -2.281 | 0.0086 | No | ||
| 73 | CIB3 | 14758 | -2.282 | 0.0150 | No | ||
| 74 | EEF1A2 | 15170 | -2.549 | -0.0001 | No | ||
| 75 | RTN4RL2 | 15225 | -2.586 | 0.0042 | No | ||
| 76 | SFTPC | 15253 | -2.605 | 0.0099 | No | ||
| 77 | MAG | 15560 | -2.842 | 0.0013 | No | ||
| 78 | KCNS1 | 15924 | -3.216 | -0.0093 | No | ||
| 79 | TRIB2 | 15938 | -3.234 | -0.0010 | No | ||
| 80 | HOXB2 | 16004 | -3.298 | 0.0047 | No | ||
| 81 | SERPINI1 | 16587 | -4.143 | -0.0152 | No | ||
| 82 | KCNA1 | 16665 | -4.283 | -0.0075 | No | ||
| 83 | BRUNOL4 | 16673 | -4.308 | 0.0042 | No | ||
| 84 | GATA4 | 17239 | -5.573 | -0.0108 | No | ||
| 85 | KDELR2 | 18169 | -9.830 | -0.0336 | No | ||
| 86 | NR2F6 | 18225 | -10.223 | -0.0081 | No | ||
| 87 | GART | 18258 | -10.491 | 0.0193 | No |