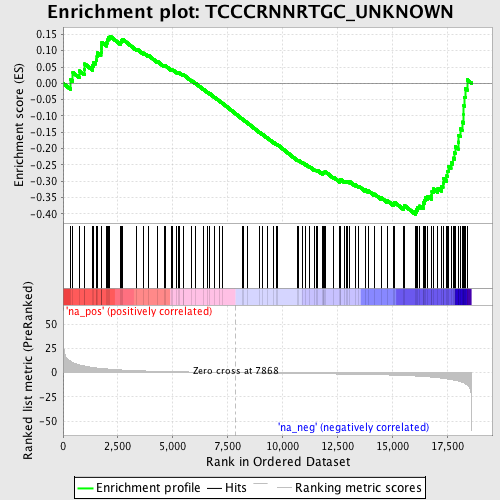
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TCCCRNNRTGC_UNKNOWN |
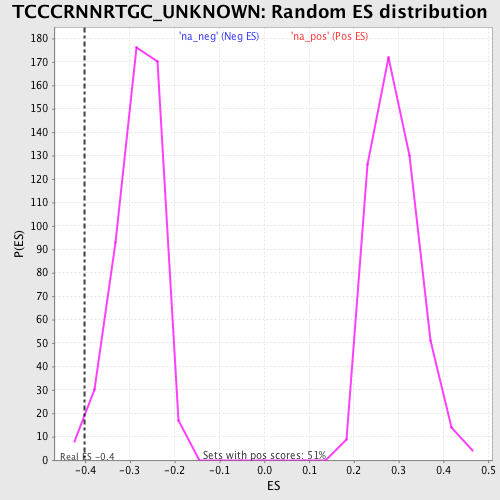
| Enrichment Score (ES) | -0.40220213 |
| Normalized Enrichment Score (NES) | -1.4231561 |
| Nominal p-value | 0.016194332 |
| FDR q-value | 0.2335788 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TLE4 | 336 | 11.842 | 0.0120 | No | ||
| 2 | INVS | 445 | 10.521 | 0.0330 | No | ||
| 3 | VCL | 736 | 8.130 | 0.0381 | No | ||
| 4 | SLC4A2 | 965 | 6.895 | 0.0433 | No | ||
| 5 | TCTA | 988 | 6.788 | 0.0595 | No | ||
| 6 | MBP | 1348 | 5.371 | 0.0538 | No | ||
| 7 | ZCCHC8 | 1398 | 5.228 | 0.0644 | No | ||
| 8 | PNRC2 | 1498 | 4.960 | 0.0717 | No | ||
| 9 | PLAGL2 | 1542 | 4.836 | 0.0817 | No | ||
| 10 | SNX17 | 1554 | 4.804 | 0.0934 | No | ||
| 11 | NSD1 | 1727 | 4.394 | 0.0953 | No | ||
| 12 | IRF2BP1 | 1739 | 4.355 | 0.1058 | No | ||
| 13 | POLK | 1753 | 4.326 | 0.1161 | No | ||
| 14 | PCTK1 | 1765 | 4.293 | 0.1265 | No | ||
| 15 | USP15 | 1993 | 3.836 | 0.1240 | No | ||
| 16 | NAP1L3 | 2005 | 3.809 | 0.1331 | No | ||
| 17 | CHD4 | 2049 | 3.732 | 0.1403 | No | ||
| 18 | CDK5 | 2136 | 3.562 | 0.1447 | No | ||
| 19 | CNOT4 | 2608 | 2.855 | 0.1266 | No | ||
| 20 | FYCO1 | 2645 | 2.801 | 0.1318 | No | ||
| 21 | FBS1 | 2715 | 2.714 | 0.1350 | No | ||
| 22 | WTAP | 3364 | 2.049 | 0.1052 | No | ||
| 23 | RBM6 | 3673 | 1.798 | 0.0931 | No | ||
| 24 | RAB1A | 3872 | 1.657 | 0.0866 | No | ||
| 25 | SNX11 | 4297 | 1.408 | 0.0673 | No | ||
| 26 | POFUT1 | 4606 | 1.218 | 0.0538 | No | ||
| 27 | PHACTR3 | 4654 | 1.198 | 0.0543 | No | ||
| 28 | DQX1 | 4948 | 1.062 | 0.0411 | No | ||
| 29 | BRMS1 | 4998 | 1.040 | 0.0411 | No | ||
| 30 | MRPL24 | 5178 | 0.957 | 0.0339 | No | ||
| 31 | SON | 5246 | 0.921 | 0.0326 | No | ||
| 32 | PLK1 | 5303 | 0.896 | 0.0319 | No | ||
| 33 | RPL7 | 5478 | 0.827 | 0.0246 | No | ||
| 34 | NUDT13 | 5487 | 0.822 | 0.0263 | No | ||
| 35 | RPS27A | 5857 | 0.683 | 0.0081 | No | ||
| 36 | MOSPD2 | 6034 | 0.609 | 0.0001 | No | ||
| 37 | TUBGCP3 | 6390 | 0.474 | -0.0179 | No | ||
| 38 | NEK2 | 6585 | 0.407 | -0.0273 | No | ||
| 39 | CASC3 | 6692 | 0.369 | -0.0321 | No | ||
| 40 | EYA4 | 6907 | 0.295 | -0.0429 | No | ||
| 41 | TRAF3IP1 | 7106 | 0.231 | -0.0530 | No | ||
| 42 | KLK9 | 7275 | 0.175 | -0.0617 | No | ||
| 43 | BET1 | 8167 | -0.093 | -0.1096 | No | ||
| 44 | BUB1B | 8240 | -0.115 | -0.1132 | No | ||
| 45 | TIMP4 | 8397 | -0.161 | -0.1212 | No | ||
| 46 | ZIC5 | 8945 | -0.329 | -0.1499 | No | ||
| 47 | SFRS1 | 9103 | -0.371 | -0.1575 | No | ||
| 48 | ASXL2 | 9300 | -0.428 | -0.1670 | No | ||
| 49 | RAD50 | 9604 | -0.516 | -0.1820 | No | ||
| 50 | BSN | 9715 | -0.547 | -0.1866 | No | ||
| 51 | RHOQ | 9778 | -0.566 | -0.1885 | No | ||
| 52 | ORC4L | 10682 | -0.819 | -0.2352 | No | ||
| 53 | RHOA | 10740 | -0.836 | -0.2362 | No | ||
| 54 | COL4A3BP | 10926 | -0.884 | -0.2439 | No | ||
| 55 | NOS3 | 11056 | -0.918 | -0.2485 | No | ||
| 56 | MORF4L1 | 11228 | -0.966 | -0.2553 | No | ||
| 57 | PUM1 | 11469 | -1.036 | -0.2656 | No | ||
| 58 | KIF11 | 11530 | -1.053 | -0.2662 | No | ||
| 59 | STMN1 | 11601 | -1.074 | -0.2672 | No | ||
| 60 | COX4I1 | 11838 | -1.149 | -0.2771 | No | ||
| 61 | IPO7 | 11849 | -1.155 | -0.2747 | No | ||
| 62 | BRS3 | 11880 | -1.162 | -0.2733 | No | ||
| 63 | RB1CC1 | 11909 | -1.169 | -0.2719 | No | ||
| 64 | PES1 | 11960 | -1.186 | -0.2715 | No | ||
| 65 | GRIN2B | 12319 | -1.292 | -0.2876 | No | ||
| 66 | PRCC | 12614 | -1.379 | -0.3000 | No | ||
| 67 | CHCHD3 | 12639 | -1.390 | -0.2977 | No | ||
| 68 | INSRR | 12650 | -1.395 | -0.2947 | No | ||
| 69 | RASSF6 | 12816 | -1.448 | -0.2999 | No | ||
| 70 | FUK | 12899 | -1.477 | -0.3006 | No | ||
| 71 | CKAP4 | 12961 | -1.498 | -0.3001 | No | ||
| 72 | PSMD11 | 13068 | -1.528 | -0.3019 | No | ||
| 73 | SFRS9 | 13314 | -1.619 | -0.3110 | No | ||
| 74 | PCM1 | 13467 | -1.679 | -0.3149 | No | ||
| 75 | UBL3 | 13759 | -1.793 | -0.3261 | No | ||
| 76 | GSK3A | 13913 | -1.860 | -0.3296 | No | ||
| 77 | LIG1 | 14187 | -1.982 | -0.3393 | No | ||
| 78 | SNX16 | 14498 | -2.141 | -0.3506 | No | ||
| 79 | SMPD3 | 14777 | -2.296 | -0.3598 | No | ||
| 80 | HDAC8 | 15056 | -2.461 | -0.3685 | No | ||
| 81 | VAPA | 15108 | -2.498 | -0.3649 | No | ||
| 82 | LZTR1 | 15523 | -2.805 | -0.3802 | No | ||
| 83 | MAG | 15560 | -2.842 | -0.3749 | No | ||
| 84 | RDH10 | 16067 | -3.376 | -0.3936 | Yes | ||
| 85 | CSNK2A1 | 16104 | -3.416 | -0.3868 | Yes | ||
| 86 | ISL1 | 16148 | -3.468 | -0.3803 | Yes | ||
| 87 | NMT1 | 16241 | -3.602 | -0.3761 | Yes | ||
| 88 | RPL30 | 16413 | -3.859 | -0.3755 | Yes | ||
| 89 | NUTF2 | 16418 | -3.866 | -0.3659 | Yes | ||
| 90 | TRIM23 | 16467 | -3.950 | -0.3584 | Yes | ||
| 91 | TCEB1 | 16512 | -4.017 | -0.3505 | Yes | ||
| 92 | RRS1 | 16620 | -4.195 | -0.3456 | Yes | ||
| 93 | VKORC1 | 16801 | -4.569 | -0.3437 | Yes | ||
| 94 | GCN5L2 | 16809 | -4.588 | -0.3324 | Yes | ||
| 95 | ACADVL | 16871 | -4.709 | -0.3236 | Yes | ||
| 96 | USP16 | 17082 | -5.147 | -0.3219 | Yes | ||
| 97 | GSPT1 | 17244 | -5.577 | -0.3164 | Yes | ||
| 98 | PA2G4 | 17316 | -5.750 | -0.3055 | Yes | ||
| 99 | HSPH1 | 17324 | -5.763 | -0.2912 | Yes | ||
| 100 | PPP1R8 | 17490 | -6.340 | -0.2840 | Yes | ||
| 101 | KPNB1 | 17532 | -6.478 | -0.2697 | Yes | ||
| 102 | TTC1 | 17561 | -6.564 | -0.2544 | Yes | ||
| 103 | MRPL50 | 17681 | -7.030 | -0.2429 | Yes | ||
| 104 | SCAMP4 | 17770 | -7.366 | -0.2289 | Yes | ||
| 105 | RPP30 | 17842 | -7.728 | -0.2130 | Yes | ||
| 106 | ABCB6 | 17872 | -7.856 | -0.1946 | Yes | ||
| 107 | IFRD2 | 18005 | -8.562 | -0.1799 | Yes | ||
| 108 | UBADC1 | 18023 | -8.674 | -0.1587 | Yes | ||
| 109 | EIF5A | 18110 | -9.263 | -0.1397 | Yes | ||
| 110 | XPOT | 18187 | -9.925 | -0.1185 | Yes | ||
| 111 | GART | 18258 | -10.491 | -0.0955 | Yes | ||
| 112 | MIR16 | 18263 | -10.525 | -0.0689 | Yes | ||
| 113 | EIF2B4 | 18287 | -10.786 | -0.0427 | Yes | ||
| 114 | PMM2 | 18331 | -11.273 | -0.0162 | Yes | ||
| 115 | POLR1A | 18417 | -12.389 | 0.0108 | Yes |