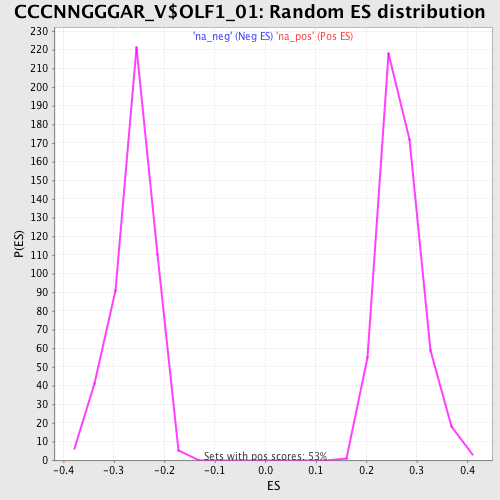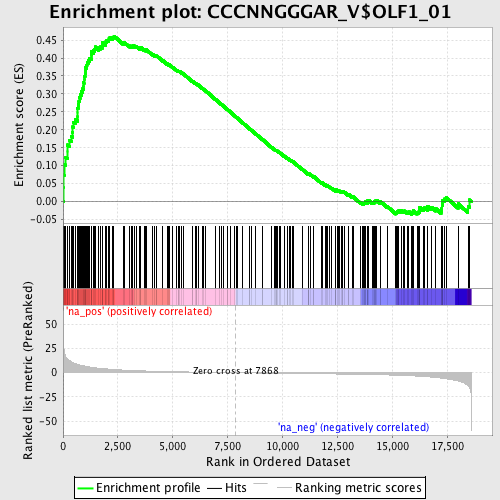
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CCCNNGGGAR_V$OLF1_01 |
| Enrichment Score (ES) | 0.46119764 |
| Normalized Enrichment Score (NES) | 1.7203189 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03996776 |
| FWER p-Value | 0.374 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABR | 21 | 28.176 | 0.0383 | Yes | ||
| 2 | ROBO3 | 27 | 25.869 | 0.0742 | Yes | ||
| 3 | CA2 | 47 | 22.087 | 0.1041 | Yes | ||
| 4 | BCL9L | 121 | 16.738 | 0.1236 | Yes | ||
| 5 | ELK3 | 205 | 13.852 | 0.1385 | Yes | ||
| 6 | LASP1 | 206 | 13.841 | 0.1578 | Yes | ||
| 7 | REPIN1 | 293 | 12.412 | 0.1705 | Yes | ||
| 8 | ETV6 | 375 | 11.290 | 0.1819 | Yes | ||
| 9 | TIAM1 | 431 | 10.657 | 0.1939 | Yes | ||
| 10 | MAGED2 | 443 | 10.526 | 0.2080 | Yes | ||
| 11 | SLC30A5 | 479 | 10.125 | 0.2203 | Yes | ||
| 12 | H1F0 | 551 | 9.387 | 0.2295 | Yes | ||
| 13 | THRA | 657 | 8.562 | 0.2358 | Yes | ||
| 14 | MESDC1 | 661 | 8.546 | 0.2476 | Yes | ||
| 15 | DLX1 | 662 | 8.535 | 0.2596 | Yes | ||
| 16 | UBAP1 | 686 | 8.408 | 0.2701 | Yes | ||
| 17 | SP4 | 719 | 8.253 | 0.2799 | Yes | ||
| 18 | TRAF4 | 740 | 8.116 | 0.2902 | Yes | ||
| 19 | FOXJ2 | 775 | 7.864 | 0.2993 | Yes | ||
| 20 | FBXL19 | 838 | 7.530 | 0.3065 | Yes | ||
| 21 | PCYT1B | 887 | 7.239 | 0.3140 | Yes | ||
| 22 | EGR1 | 929 | 7.104 | 0.3217 | Yes | ||
| 23 | H3F3B | 935 | 7.045 | 0.3313 | Yes | ||
| 24 | ICA1 | 956 | 6.935 | 0.3400 | Yes | ||
| 25 | BMP4 | 961 | 6.913 | 0.3494 | Yes | ||
| 26 | GNB2 | 1001 | 6.736 | 0.3567 | Yes | ||
| 27 | TRIM8 | 1020 | 6.649 | 0.3651 | Yes | ||
| 28 | ABI3 | 1032 | 6.597 | 0.3737 | Yes | ||
| 29 | DTX3 | 1077 | 6.436 | 0.3803 | Yes | ||
| 30 | TNIP1 | 1090 | 6.378 | 0.3886 | Yes | ||
| 31 | AXL | 1143 | 6.168 | 0.3944 | Yes | ||
| 32 | ZDHHC14 | 1203 | 5.904 | 0.3995 | Yes | ||
| 33 | BAZ2A | 1283 | 5.590 | 0.4030 | Yes | ||
| 34 | ICAM1 | 1285 | 5.589 | 0.4108 | Yes | ||
| 35 | HIC2 | 1301 | 5.527 | 0.4177 | Yes | ||
| 36 | RHOG | 1387 | 5.256 | 0.4204 | Yes | ||
| 37 | MADCAM1 | 1444 | 5.104 | 0.4245 | Yes | ||
| 38 | TST | 1454 | 5.080 | 0.4311 | Yes | ||
| 39 | LIMK2 | 1627 | 4.618 | 0.4283 | Yes | ||
| 40 | TMEM24 | 1688 | 4.469 | 0.4313 | Yes | ||
| 41 | ATF5 | 1778 | 4.272 | 0.4324 | Yes | ||
| 42 | PIAS1 | 1786 | 4.255 | 0.4380 | Yes | ||
| 43 | FCHSD2 | 1797 | 4.240 | 0.4434 | Yes | ||
| 44 | PPP3CB | 1943 | 3.938 | 0.4410 | Yes | ||
| 45 | ARMCX3 | 1952 | 3.919 | 0.4461 | Yes | ||
| 46 | PSCD3 | 1991 | 3.837 | 0.4494 | Yes | ||
| 47 | MAPKAP1 | 2073 | 3.682 | 0.4501 | Yes | ||
| 48 | AKAP12 | 2090 | 3.653 | 0.4544 | Yes | ||
| 49 | PACSIN1 | 2134 | 3.572 | 0.4570 | Yes | ||
| 50 | ARID1A | 2251 | 3.402 | 0.4555 | Yes | ||
| 51 | TNKS1BP1 | 2282 | 3.333 | 0.4585 | Yes | ||
| 52 | BAT2 | 2318 | 3.271 | 0.4612 | Yes | ||
| 53 | PLAG1 | 2740 | 2.688 | 0.4421 | No | ||
| 54 | DUSP4 | 2783 | 2.646 | 0.4435 | No | ||
| 55 | POU2AF1 | 3020 | 2.376 | 0.4340 | No | ||
| 56 | MPST | 3125 | 2.277 | 0.4316 | No | ||
| 57 | IL11 | 3166 | 2.229 | 0.4325 | No | ||
| 58 | CD79B | 3181 | 2.216 | 0.4349 | No | ||
| 59 | ADAMTS4 | 3257 | 2.139 | 0.4338 | No | ||
| 60 | LUC7L2 | 3322 | 2.088 | 0.4332 | No | ||
| 61 | HEBP2 | 3501 | 1.934 | 0.4263 | No | ||
| 62 | LHX2 | 3538 | 1.909 | 0.4270 | No | ||
| 63 | HCST | 3543 | 1.904 | 0.4294 | No | ||
| 64 | WDR13 | 3720 | 1.770 | 0.4223 | No | ||
| 65 | GRM7 | 3772 | 1.730 | 0.4220 | No | ||
| 66 | COL23A1 | 3793 | 1.714 | 0.4233 | No | ||
| 67 | PTMA | 4071 | 1.542 | 0.4104 | No | ||
| 68 | BCOR | 4172 | 1.490 | 0.4071 | No | ||
| 69 | CLDN15 | 4248 | 1.436 | 0.4050 | No | ||
| 70 | RNF44 | 4265 | 1.429 | 0.4061 | No | ||
| 71 | NDFIP1 | 4541 | 1.255 | 0.3930 | No | ||
| 72 | GRIK1 | 4735 | 1.164 | 0.3841 | No | ||
| 73 | JMJD2A | 4816 | 1.122 | 0.3813 | No | ||
| 74 | JMJD1B | 4845 | 1.109 | 0.3814 | No | ||
| 75 | SSH2 | 5006 | 1.035 | 0.3741 | No | ||
| 76 | SYTL2 | 5169 | 0.962 | 0.3667 | No | ||
| 77 | SNCB | 5259 | 0.915 | 0.3631 | No | ||
| 78 | SLC9A6 | 5278 | 0.908 | 0.3634 | No | ||
| 79 | VAT1 | 5297 | 0.897 | 0.3637 | No | ||
| 80 | FKBP2 | 5415 | 0.853 | 0.3585 | No | ||
| 81 | EFS | 5468 | 0.830 | 0.3569 | No | ||
| 82 | MAGEL2 | 5888 | 0.669 | 0.3350 | No | ||
| 83 | SP3 | 5919 | 0.660 | 0.3343 | No | ||
| 84 | LIN28 | 6013 | 0.622 | 0.3302 | No | ||
| 85 | LPP | 6075 | 0.590 | 0.3277 | No | ||
| 86 | HOXA11 | 6086 | 0.585 | 0.3279 | No | ||
| 87 | HPCA | 6152 | 0.562 | 0.3252 | No | ||
| 88 | DLL4 | 6355 | 0.486 | 0.3149 | No | ||
| 89 | NF2 | 6410 | 0.468 | 0.3126 | No | ||
| 90 | CHAT | 6484 | 0.439 | 0.3093 | No | ||
| 91 | UBTF | 6961 | 0.280 | 0.2838 | No | ||
| 92 | CDON | 7142 | 0.220 | 0.2744 | No | ||
| 93 | PIGW | 7203 | 0.200 | 0.2714 | No | ||
| 94 | SLC41A1 | 7321 | 0.161 | 0.2652 | No | ||
| 95 | IL4I1 | 7481 | 0.117 | 0.2568 | No | ||
| 96 | IGF2R | 7489 | 0.114 | 0.2566 | No | ||
| 97 | MLLT6 | 7613 | 0.078 | 0.2500 | No | ||
| 98 | CD72 | 7788 | 0.022 | 0.2406 | No | ||
| 99 | ENPP2 | 7917 | -0.013 | 0.2336 | No | ||
| 100 | C1QTNF1 | 7962 | -0.028 | 0.2313 | No | ||
| 101 | APBA1 | 8194 | -0.101 | 0.2189 | No | ||
| 102 | TNNT3 | 8475 | -0.184 | 0.2039 | No | ||
| 103 | AAMP | 8516 | -0.198 | 0.2020 | No | ||
| 104 | ANXA9 | 8592 | -0.220 | 0.1983 | No | ||
| 105 | NEUROD2 | 8747 | -0.268 | 0.1903 | No | ||
| 106 | GNAO1 | 9081 | -0.365 | 0.1727 | No | ||
| 107 | ATP2B3 | 9104 | -0.371 | 0.1720 | No | ||
| 108 | ELL2 | 9518 | -0.491 | 0.1503 | No | ||
| 109 | STAG2 | 9636 | -0.524 | 0.1447 | No | ||
| 110 | ECM2 | 9692 | -0.541 | 0.1424 | No | ||
| 111 | WNT7B | 9707 | -0.546 | 0.1424 | No | ||
| 112 | TBX21 | 9726 | -0.550 | 0.1422 | No | ||
| 113 | RHOQ | 9778 | -0.566 | 0.1402 | No | ||
| 114 | ADRB1 | 9865 | -0.590 | 0.1364 | No | ||
| 115 | GREM1 | 9928 | -0.609 | 0.1339 | No | ||
| 116 | RORC | 10073 | -0.650 | 0.1270 | No | ||
| 117 | GJC1 | 10219 | -0.689 | 0.1201 | No | ||
| 118 | TTYH1 | 10220 | -0.689 | 0.1210 | No | ||
| 119 | ETV4 | 10337 | -0.720 | 0.1157 | No | ||
| 120 | CHL1 | 10372 | -0.728 | 0.1149 | No | ||
| 121 | LRRTM3 | 10445 | -0.749 | 0.1120 | No | ||
| 122 | JARID2 | 10491 | -0.764 | 0.1107 | No | ||
| 123 | FGF11 | 10908 | -0.880 | 0.0893 | No | ||
| 124 | ALOX12B | 11161 | -0.948 | 0.0770 | No | ||
| 125 | EVI1 | 11169 | -0.951 | 0.0779 | No | ||
| 126 | PRRX1 | 11202 | -0.958 | 0.0775 | No | ||
| 127 | MDGA1 | 11291 | -0.985 | 0.0741 | No | ||
| 128 | SGK2 | 11418 | -1.025 | 0.0687 | No | ||
| 129 | GRK5 | 11433 | -1.029 | 0.0694 | No | ||
| 130 | PDGFC | 11771 | -1.129 | 0.0527 | No | ||
| 131 | CNTN2 | 11799 | -1.137 | 0.0528 | No | ||
| 132 | TOM1L1 | 11941 | -1.179 | 0.0468 | No | ||
| 133 | ADCYAP1 | 12020 | -1.204 | 0.0442 | No | ||
| 134 | LRFN3 | 12067 | -1.218 | 0.0434 | No | ||
| 135 | PTHLH | 12160 | -1.248 | 0.0402 | No | ||
| 136 | FSTL3 | 12240 | -1.270 | 0.0377 | No | ||
| 137 | HOXA6 | 12409 | -1.321 | 0.0304 | No | ||
| 138 | EPHB3 | 12426 | -1.327 | 0.0314 | No | ||
| 139 | BAI2 | 12429 | -1.327 | 0.0331 | No | ||
| 140 | CALM1 | 12515 | -1.352 | 0.0304 | No | ||
| 141 | LRP8 | 12534 | -1.356 | 0.0313 | No | ||
| 142 | BANP | 12603 | -1.376 | 0.0296 | No | ||
| 143 | HOXC10 | 12681 | -1.404 | 0.0273 | No | ||
| 144 | LRRC20 | 12721 | -1.418 | 0.0272 | No | ||
| 145 | NFKBIA | 12722 | -1.418 | 0.0292 | No | ||
| 146 | SLC25A28 | 12817 | -1.449 | 0.0261 | No | ||
| 147 | TRIM29 | 13005 | -1.510 | 0.0181 | No | ||
| 148 | MYOZ1 | 13028 | -1.516 | 0.0190 | No | ||
| 149 | ADCY4 | 13175 | -1.566 | 0.0133 | No | ||
| 150 | KCND1 | 13220 | -1.582 | 0.0131 | No | ||
| 151 | GRIN2D | 13561 | -1.718 | -0.0030 | No | ||
| 152 | NUP62 | 13644 | -1.743 | -0.0050 | No | ||
| 153 | GNGT2 | 13708 | -1.773 | -0.0059 | No | ||
| 154 | FLT1 | 13723 | -1.780 | -0.0042 | No | ||
| 155 | UPF3B | 13757 | -1.792 | -0.0035 | No | ||
| 156 | SDCCAG8 | 13760 | -1.795 | -0.0011 | No | ||
| 157 | PAX2 | 13775 | -1.802 | 0.0007 | No | ||
| 158 | POGK | 13879 | -1.845 | -0.0023 | No | ||
| 159 | CKM | 13880 | -1.845 | 0.0002 | No | ||
| 160 | C1QL1 | 13888 | -1.847 | 0.0025 | No | ||
| 161 | CHCHD7 | 13938 | -1.871 | 0.0024 | No | ||
| 162 | CHRDL1 | 14098 | -1.940 | -0.0035 | No | ||
| 163 | RBM10 | 14146 | -1.965 | -0.0033 | No | ||
| 164 | LRP1 | 14171 | -1.976 | -0.0019 | No | ||
| 165 | THBS3 | 14184 | -1.980 | 0.0003 | No | ||
| 166 | RAB10 | 14219 | -1.996 | 0.0012 | No | ||
| 167 | ELAVL3 | 14255 | -2.015 | 0.0021 | No | ||
| 168 | DDR1 | 14293 | -2.036 | 0.0030 | No | ||
| 169 | GRIK3 | 14443 | -2.113 | -0.0022 | No | ||
| 170 | HR | 14469 | -2.130 | -0.0005 | No | ||
| 171 | SMPD3 | 14777 | -2.296 | -0.0140 | No | ||
| 172 | GPM6B | 15162 | -2.539 | -0.0313 | No | ||
| 173 | DAB1 | 15204 | -2.570 | -0.0299 | No | ||
| 174 | COL18A1 | 15242 | -2.597 | -0.0283 | No | ||
| 175 | PLXDC2 | 15279 | -2.631 | -0.0266 | No | ||
| 176 | MTSS1 | 15304 | -2.646 | -0.0242 | No | ||
| 177 | DPF1 | 15406 | -2.724 | -0.0259 | No | ||
| 178 | COL11A2 | 15507 | -2.794 | -0.0274 | No | ||
| 179 | MAG | 15560 | -2.842 | -0.0262 | No | ||
| 180 | SFRP2 | 15706 | -2.965 | -0.0299 | No | ||
| 181 | SCG3 | 15761 | -3.028 | -0.0286 | No | ||
| 182 | UBE2N | 15900 | -3.182 | -0.0317 | No | ||
| 183 | TRIB2 | 15938 | -3.234 | -0.0292 | No | ||
| 184 | VASP | 15953 | -3.246 | -0.0254 | No | ||
| 185 | SLC25A14 | 16130 | -3.443 | -0.0301 | No | ||
| 186 | STC2 | 16204 | -3.551 | -0.0291 | No | ||
| 187 | EPN3 | 16240 | -3.601 | -0.0260 | No | ||
| 188 | NCDN | 16251 | -3.608 | -0.0215 | No | ||
| 189 | RGS3 | 16253 | -3.614 | -0.0165 | No | ||
| 190 | PCBP4 | 16424 | -3.874 | -0.0203 | No | ||
| 191 | NR2E1 | 16480 | -3.972 | -0.0177 | No | ||
| 192 | NEO1 | 16603 | -4.176 | -0.0185 | No | ||
| 193 | TPM1 | 16613 | -4.188 | -0.0131 | No | ||
| 194 | TENC1 | 16791 | -4.556 | -0.0164 | No | ||
| 195 | MYOHD1 | 16980 | -4.895 | -0.0197 | No | ||
| 196 | ATP1A1 | 17226 | -5.543 | -0.0253 | No | ||
| 197 | NDUFS2 | 17262 | -5.625 | -0.0193 | No | ||
| 198 | PDAP1 | 17268 | -5.641 | -0.0117 | No | ||
| 199 | DPYSL5 | 17279 | -5.667 | -0.0043 | No | ||
| 200 | PSME3 | 17304 | -5.719 | 0.0024 | No | ||
| 201 | SMARCE1 | 17371 | -5.927 | 0.0071 | No | ||
| 202 | DUSP22 | 17467 | -6.214 | 0.0107 | No | ||
| 203 | RAB39 | 18001 | -8.541 | -0.0063 | No | ||
| 204 | MTX1 | 18453 | -13.048 | -0.0126 | No | ||
| 205 | GCNT2 | 18533 | -15.255 | 0.0045 | No |