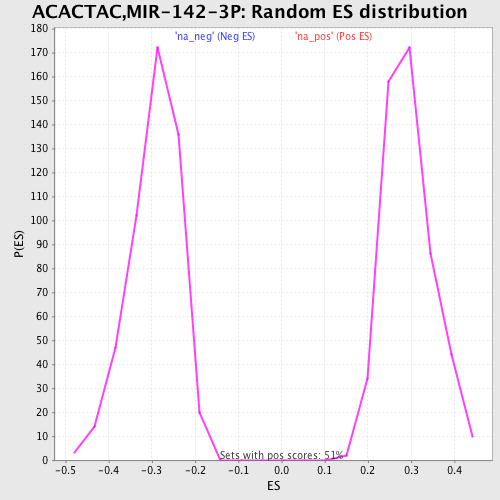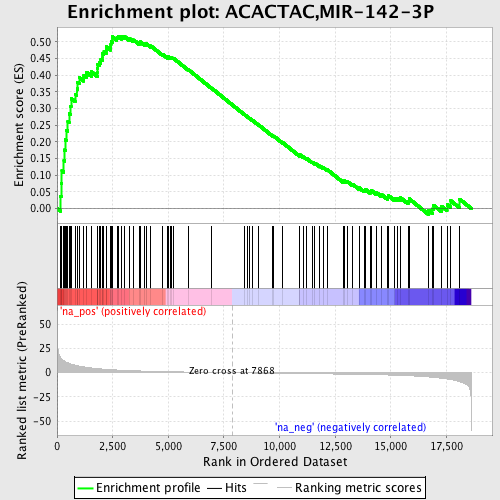
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ACACTAC,MIR-142-3P |
| Enrichment Score (ES) | 0.51733136 |
| Normalized Enrichment Score (NES) | 1.7640034 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.033359263 |
| FWER p-Value | 0.242 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SGK | 143 | 15.627 | 0.0371 | Yes | ||
| 2 | MARCKS | 184 | 14.244 | 0.0758 | Yes | ||
| 3 | FMNL2 | 217 | 13.686 | 0.1134 | Yes | ||
| 4 | PRLR | 307 | 12.224 | 0.1436 | Yes | ||
| 5 | BACH1 | 338 | 11.788 | 0.1758 | Yes | ||
| 6 | MAP3K11 | 373 | 11.326 | 0.2065 | Yes | ||
| 7 | ERG | 426 | 10.702 | 0.2344 | Yes | ||
| 8 | ARNTL | 485 | 10.085 | 0.2602 | Yes | ||
| 9 | ATF7IP | 541 | 9.500 | 0.2845 | Yes | ||
| 10 | STAU1 | 612 | 8.898 | 0.3062 | Yes | ||
| 11 | MAP3K7IP2 | 630 | 8.758 | 0.3304 | Yes | ||
| 12 | PREI3 | 812 | 7.658 | 0.3426 | Yes | ||
| 13 | TGFBR1 | 893 | 7.218 | 0.3590 | Yes | ||
| 14 | GNAQ | 918 | 7.135 | 0.3782 | Yes | ||
| 15 | GNB2 | 1001 | 6.736 | 0.3931 | Yes | ||
| 16 | STAM | 1201 | 5.908 | 0.3993 | Yes | ||
| 17 | RNF31 | 1314 | 5.480 | 0.4090 | Yes | ||
| 18 | STRN3 | 1544 | 4.832 | 0.4105 | Yes | ||
| 19 | PDE4B | 1805 | 4.224 | 0.4086 | Yes | ||
| 20 | PCAF | 1812 | 4.215 | 0.4203 | Yes | ||
| 21 | ADCY9 | 1827 | 4.185 | 0.4316 | Yes | ||
| 22 | INPP5A | 1899 | 4.022 | 0.4393 | Yes | ||
| 23 | PTPN23 | 1966 | 3.892 | 0.4469 | Yes | ||
| 24 | GFI1 | 2036 | 3.762 | 0.4540 | Yes | ||
| 25 | ITGAV | 2042 | 3.752 | 0.4645 | Yes | ||
| 26 | PURB | 2106 | 3.628 | 0.4715 | Yes | ||
| 27 | RGL2 | 2205 | 3.479 | 0.4762 | Yes | ||
| 28 | ASB7 | 2222 | 3.461 | 0.4852 | Yes | ||
| 29 | SNF1LK2 | 2405 | 3.135 | 0.4844 | Yes | ||
| 30 | APC | 2416 | 3.120 | 0.4928 | Yes | ||
| 31 | ROCK2 | 2424 | 3.107 | 0.5014 | Yes | ||
| 32 | CFL2 | 2477 | 3.045 | 0.5073 | Yes | ||
| 33 | UTX | 2480 | 3.038 | 0.5159 | Yes | ||
| 34 | VAMP3 | 2691 | 2.745 | 0.5124 | Yes | ||
| 35 | FBXO3 | 2744 | 2.686 | 0.5173 | Yes | ||
| 36 | MGAT4A | 2914 | 2.487 | 0.5153 | No | ||
| 37 | RAB40C | 3023 | 2.372 | 0.5163 | No | ||
| 38 | ACVR2A | 3262 | 2.133 | 0.5096 | No | ||
| 39 | LRRC1 | 3438 | 1.978 | 0.5058 | No | ||
| 40 | MMD | 3688 | 1.788 | 0.4975 | No | ||
| 41 | ATG16L1 | 3733 | 1.761 | 0.5002 | No | ||
| 42 | HMGB1 | 3937 | 1.617 | 0.4939 | No | ||
| 43 | RERE | 4000 | 1.577 | 0.4950 | No | ||
| 44 | CCNT2 | 4208 | 1.462 | 0.4881 | No | ||
| 45 | UTY | 4742 | 1.160 | 0.4626 | No | ||
| 46 | EML4 | 4975 | 1.048 | 0.4531 | No | ||
| 47 | PPFIA1 | 4987 | 1.044 | 0.4555 | No | ||
| 48 | FKBP1A | 5087 | 1.000 | 0.4530 | No | ||
| 49 | MYST2 | 5151 | 0.970 | 0.4524 | No | ||
| 50 | RAC1 | 5218 | 0.936 | 0.4515 | No | ||
| 51 | TIPARP | 5895 | 0.667 | 0.4169 | No | ||
| 52 | EDG3 | 6940 | 0.287 | 0.3614 | No | ||
| 53 | SLC7A11 | 8417 | -0.167 | 0.2822 | No | ||
| 54 | EGR2 | 8574 | -0.213 | 0.2744 | No | ||
| 55 | ARL15 | 8641 | -0.237 | 0.2715 | No | ||
| 56 | EHF | 8765 | -0.276 | 0.2656 | No | ||
| 57 | BACH2 | 9069 | -0.361 | 0.2503 | No | ||
| 58 | STX12 | 9689 | -0.539 | 0.2185 | No | ||
| 59 | MLLT7 | 9747 | -0.557 | 0.2170 | No | ||
| 60 | BTBD7 | 10138 | -0.666 | 0.1978 | No | ||
| 61 | ATP2A2 | 10896 | -0.876 | 0.1595 | No | ||
| 62 | CLTA | 10912 | -0.881 | 0.1612 | No | ||
| 63 | BCLAF1 | 11077 | -0.924 | 0.1550 | No | ||
| 64 | MORF4L1 | 11228 | -0.966 | 0.1497 | No | ||
| 65 | PUM1 | 11469 | -1.036 | 0.1397 | No | ||
| 66 | XPO1 | 11588 | -1.069 | 0.1364 | No | ||
| 67 | ARID5B | 11806 | -1.140 | 0.1279 | No | ||
| 68 | AFF2 | 11969 | -1.191 | 0.1226 | No | ||
| 69 | PSIP1 | 12132 | -1.240 | 0.1174 | No | ||
| 70 | FNDC3A | 12858 | -1.463 | 0.0825 | No | ||
| 71 | COPG | 12925 | -1.486 | 0.0832 | No | ||
| 72 | CPEB2 | 13043 | -1.520 | 0.0812 | No | ||
| 73 | SNF1LK | 13289 | -1.610 | 0.0726 | No | ||
| 74 | SLCO4C1 | 13570 | -1.721 | 0.0624 | No | ||
| 75 | RARG | 13800 | -1.812 | 0.0553 | No | ||
| 76 | HMGA2 | 13856 | -1.835 | 0.0576 | No | ||
| 77 | SP8 | 14068 | -1.927 | 0.0517 | No | ||
| 78 | SOX11 | 14132 | -1.958 | 0.0539 | No | ||
| 79 | TRPS1 | 14365 | -2.071 | 0.0473 | No | ||
| 80 | IRAK1 | 14586 | -2.187 | 0.0417 | No | ||
| 81 | ACSL4 | 14853 | -2.338 | 0.0341 | No | ||
| 82 | AKT1S1 | 14898 | -2.361 | 0.0385 | No | ||
| 83 | CRTAM | 15176 | -2.552 | 0.0308 | No | ||
| 84 | SPRED1 | 15312 | -2.650 | 0.0312 | No | ||
| 85 | MORF4L2 | 15433 | -2.741 | 0.0326 | No | ||
| 86 | TIRAP | 15780 | -3.057 | 0.0226 | No | ||
| 87 | GTF2A1 | 15820 | -3.112 | 0.0295 | No | ||
| 88 | SYPL1 | 16696 | -4.355 | -0.0053 | No | ||
| 89 | CRK | 16873 | -4.709 | -0.0013 | No | ||
| 90 | CSTF3 | 16921 | -4.791 | 0.0099 | No | ||
| 91 | ANK3 | 17277 | -5.660 | 0.0070 | No | ||
| 92 | TARDBP | 17527 | -6.456 | 0.0121 | No | ||
| 93 | DIRC2 | 17697 | -7.073 | 0.0233 | No | ||
| 94 | COPS7A | 18093 | -9.164 | 0.0282 | No |