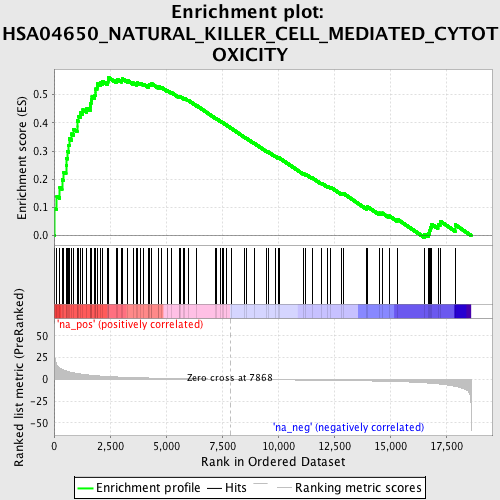
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04650_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY |
| Enrichment Score (ES) | 0.56046444 |
| Normalized Enrichment Score (NES) | 1.9265577 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05031472 |
| FWER p-Value | 0.078 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0969 | Yes | ||
| 2 | NFATC1 | 123 | 16.658 | 0.1394 | Yes | ||
| 3 | IFNAR2 | 256 | 12.952 | 0.1700 | Yes | ||
| 4 | VAV1 | 354 | 11.615 | 0.1986 | Yes | ||
| 5 | LAT | 432 | 10.644 | 0.2254 | Yes | ||
| 6 | TNFSF10 | 532 | 9.625 | 0.2481 | Yes | ||
| 7 | FYN | 553 | 9.371 | 0.2743 | Yes | ||
| 8 | RAC2 | 606 | 8.959 | 0.2976 | Yes | ||
| 9 | PIK3CG | 650 | 8.605 | 0.3203 | Yes | ||
| 10 | SH3BP2 | 676 | 8.459 | 0.3436 | Yes | ||
| 11 | LCP2 | 783 | 7.814 | 0.3606 | Yes | ||
| 12 | NFAT5 | 871 | 7.334 | 0.3772 | Yes | ||
| 13 | ZAP70 | 1027 | 6.606 | 0.3881 | Yes | ||
| 14 | PRKCB1 | 1028 | 6.604 | 0.4073 | Yes | ||
| 15 | ITGAL | 1097 | 6.349 | 0.4221 | Yes | ||
| 16 | LCK | 1176 | 6.008 | 0.4354 | Yes | ||
| 17 | ICAM1 | 1285 | 5.589 | 0.4459 | Yes | ||
| 18 | PIK3CD | 1460 | 5.066 | 0.4512 | Yes | ||
| 19 | PLCG2 | 1612 | 4.665 | 0.4566 | Yes | ||
| 20 | KLRD1 | 1629 | 4.617 | 0.4692 | Yes | ||
| 21 | PTK2B | 1651 | 4.557 | 0.4813 | Yes | ||
| 22 | MAPK3 | 1689 | 4.468 | 0.4924 | Yes | ||
| 23 | PAK1 | 1819 | 4.193 | 0.4976 | Yes | ||
| 24 | KRAS | 1847 | 4.132 | 0.5082 | Yes | ||
| 25 | PPP3CC | 1849 | 4.130 | 0.5201 | Yes | ||
| 26 | IFNGR2 | 1916 | 3.987 | 0.5282 | Yes | ||
| 27 | PPP3CB | 1943 | 3.938 | 0.5382 | Yes | ||
| 28 | SOS2 | 2078 | 3.676 | 0.5417 | Yes | ||
| 29 | GRB2 | 2166 | 3.530 | 0.5473 | Yes | ||
| 30 | ITGB2 | 2393 | 3.152 | 0.5443 | Yes | ||
| 31 | NFATC3 | 2407 | 3.130 | 0.5527 | Yes | ||
| 32 | IFNGR1 | 2431 | 3.101 | 0.5605 | Yes | ||
| 33 | PPP3CA | 2779 | 2.647 | 0.5494 | No | ||
| 34 | FCER1G | 2820 | 2.607 | 0.5549 | No | ||
| 35 | PRF1 | 3012 | 2.385 | 0.5515 | No | ||
| 36 | CSF2 | 3037 | 2.356 | 0.5571 | No | ||
| 37 | FCGR3A | 3284 | 2.114 | 0.5499 | No | ||
| 38 | HCST | 3543 | 1.904 | 0.5415 | No | ||
| 39 | CD247 | 3691 | 1.785 | 0.5388 | No | ||
| 40 | IFNAR1 | 3707 | 1.777 | 0.5432 | No | ||
| 41 | TNF | 3842 | 1.682 | 0.5408 | No | ||
| 42 | FAS | 4002 | 1.576 | 0.5368 | No | ||
| 43 | PPP3R1 | 4214 | 1.460 | 0.5297 | No | ||
| 44 | VAV2 | 4233 | 1.446 | 0.5329 | No | ||
| 45 | PIK3R1 | 4260 | 1.432 | 0.5357 | No | ||
| 46 | PIK3R5 | 4339 | 1.390 | 0.5355 | No | ||
| 47 | KIR3DL1 | 4360 | 1.370 | 0.5384 | No | ||
| 48 | CD48 | 4643 | 1.201 | 0.5267 | No | ||
| 49 | SOS1 | 4644 | 1.201 | 0.5302 | No | ||
| 50 | NCR1 | 4787 | 1.138 | 0.5259 | No | ||
| 51 | MAP2K1 | 5069 | 1.006 | 0.5136 | No | ||
| 52 | RAC1 | 5218 | 0.936 | 0.5083 | No | ||
| 53 | IFNA13 | 5587 | 0.779 | 0.4907 | No | ||
| 54 | PPP3R2 | 5594 | 0.776 | 0.4927 | No | ||
| 55 | RAC3 | 5638 | 0.760 | 0.4926 | No | ||
| 56 | RAF1 | 5794 | 0.702 | 0.4862 | No | ||
| 57 | SHC3 | 5828 | 0.691 | 0.4865 | No | ||
| 58 | IFNB1 | 5987 | 0.632 | 0.4798 | No | ||
| 59 | ARAF | 6377 | 0.479 | 0.4602 | No | ||
| 60 | CASP3 | 7195 | 0.204 | 0.4167 | No | ||
| 61 | PLCG1 | 7237 | 0.186 | 0.4150 | No | ||
| 62 | GZMB | 7414 | 0.138 | 0.4059 | No | ||
| 63 | TYROBP | 7501 | 0.112 | 0.4016 | No | ||
| 64 | PIK3CA | 7547 | 0.100 | 0.3994 | No | ||
| 65 | FASLG | 7706 | 0.045 | 0.3910 | No | ||
| 66 | KLRC3 | 7909 | -0.011 | 0.3801 | No | ||
| 67 | PRKCA | 8501 | -0.195 | 0.3488 | No | ||
| 68 | SH2D1B | 8589 | -0.219 | 0.3447 | No | ||
| 69 | KLRC2 | 8935 | -0.327 | 0.3271 | No | ||
| 70 | SH2D1A | 9472 | -0.480 | 0.2995 | No | ||
| 71 | IFNA4 | 9550 | -0.501 | 0.2968 | No | ||
| 72 | IFNA14 | 9880 | -0.596 | 0.2808 | No | ||
| 73 | IFNA5 | 10028 | -0.638 | 0.2747 | No | ||
| 74 | IFNA7 | 10043 | -0.642 | 0.2758 | No | ||
| 75 | KLRK1 | 11140 | -0.942 | 0.2194 | No | ||
| 76 | NFATC2 | 11235 | -0.968 | 0.2171 | No | ||
| 77 | IFNG | 11523 | -1.051 | 0.2047 | No | ||
| 78 | KLRC1 | 11928 | -1.175 | 0.1863 | No | ||
| 79 | NFATC4 | 12203 | -1.261 | 0.1752 | No | ||
| 80 | IFNA6 | 12359 | -1.306 | 0.1706 | No | ||
| 81 | SYK | 12848 | -1.460 | 0.1485 | No | ||
| 82 | SHC1 | 12913 | -1.483 | 0.1494 | No | ||
| 83 | IFNA2 | 13947 | -1.878 | 0.0991 | No | ||
| 84 | IFNA1 | 13979 | -1.891 | 0.1029 | No | ||
| 85 | TNFRSF10B | 14515 | -2.155 | 0.0803 | No | ||
| 86 | HRAS | 14637 | -2.213 | 0.0802 | No | ||
| 87 | MAP2K2 | 14962 | -2.404 | 0.0697 | No | ||
| 88 | PIK3R2 | 15311 | -2.649 | 0.0586 | No | ||
| 89 | BID | 16523 | -4.040 | 0.0050 | No | ||
| 90 | PTPN6 | 16706 | -4.373 | 0.0079 | No | ||
| 91 | MAPK1 | 16773 | -4.517 | 0.0175 | No | ||
| 92 | NRAS | 16794 | -4.560 | 0.0297 | No | ||
| 93 | PTPN11 | 16849 | -4.668 | 0.0404 | No | ||
| 94 | ICAM2 | 17138 | -5.317 | 0.0403 | No | ||
| 95 | PIK3R3 | 17231 | -5.558 | 0.0515 | No | ||
| 96 | PIK3CB | 17904 | -7.974 | 0.0384 | No |
