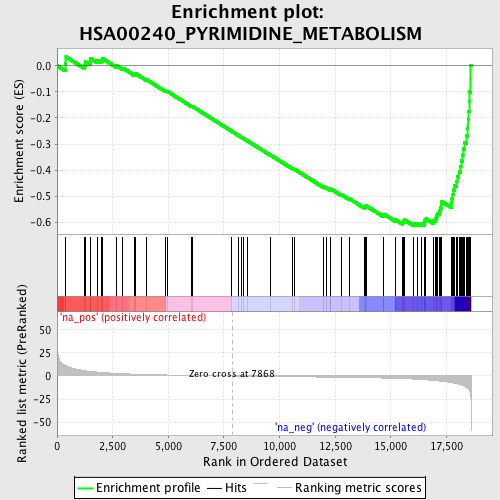
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.61336 |
| Normalized Enrichment Score (NES) | -2.07988 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DCK | 364 | 11.502 | 0.0087 | No | ||
| 2 | NP | 398 | 10.952 | 0.0340 | No | ||
| 3 | NT5M | 1239 | 5.775 | 0.0029 | No | ||
| 4 | NT5C2 | 1281 | 5.608 | 0.0145 | No | ||
| 5 | TK1 | 1490 | 4.983 | 0.0156 | No | ||
| 6 | RRM2 | 1494 | 4.971 | 0.0277 | No | ||
| 7 | POLD1 | 1806 | 4.224 | 0.0214 | No | ||
| 8 | POLD3 | 1999 | 3.822 | 0.0204 | No | ||
| 9 | ZNRD1 | 2056 | 3.714 | 0.0266 | No | ||
| 10 | TYMS | 2677 | 2.752 | -0.0001 | No | ||
| 11 | TK2 | 2956 | 2.440 | -0.0091 | No | ||
| 12 | UPP1 | 3499 | 1.935 | -0.0335 | No | ||
| 13 | POLD4 | 3542 | 1.904 | -0.0311 | No | ||
| 14 | TXNRD1 | 4032 | 1.563 | -0.0536 | No | ||
| 15 | RRM1 | 4858 | 1.103 | -0.0954 | No | ||
| 16 | ITPA | 4953 | 1.060 | -0.0978 | No | ||
| 17 | POLA1 | 6048 | 0.604 | -0.1553 | No | ||
| 18 | POLR2C | 6074 | 0.590 | -0.1552 | No | ||
| 19 | POLE2 | 7824 | 0.012 | -0.2495 | No | ||
| 20 | ENTPD3 | 8146 | -0.088 | -0.2666 | No | ||
| 21 | DPYS | 8290 | -0.129 | -0.2740 | No | ||
| 22 | CANT1 | 8383 | -0.158 | -0.2786 | No | ||
| 23 | ENTPD5 | 8571 | -0.213 | -0.2882 | No | ||
| 24 | AICDA | 9572 | -0.507 | -0.3409 | No | ||
| 25 | CTPS2 | 10571 | -0.786 | -0.3927 | No | ||
| 26 | ENTPD1 | 10687 | -0.821 | -0.3969 | No | ||
| 27 | DPYD | 11955 | -1.184 | -0.4623 | No | ||
| 28 | NUDT2 | 12088 | -1.226 | -0.4664 | No | ||
| 29 | NT5C1B | 12303 | -1.288 | -0.4748 | No | ||
| 30 | POLR2B | 12304 | -1.288 | -0.4716 | No | ||
| 31 | ENTPD8 | 12762 | -1.430 | -0.4927 | No | ||
| 32 | POLE4 | 13148 | -1.558 | -0.5097 | No | ||
| 33 | POLR2I | 13802 | -1.813 | -0.5404 | No | ||
| 34 | RRM2B | 13840 | -1.828 | -0.5379 | No | ||
| 35 | UPP2 | 13899 | -1.853 | -0.5365 | No | ||
| 36 | ENTPD6 | 14672 | -2.237 | -0.5726 | No | ||
| 37 | CDA | 14683 | -2.243 | -0.5676 | No | ||
| 38 | UPB1 | 15216 | -2.577 | -0.5899 | No | ||
| 39 | PNPT1 | 15517 | -2.804 | -0.5992 | No | ||
| 40 | PRIM1 | 15556 | -2.840 | -0.5942 | No | ||
| 41 | NT5E | 15592 | -2.872 | -0.5890 | No | ||
| 42 | ECGF1 | 16018 | -3.321 | -0.6038 | No | ||
| 43 | TXNRD2 | 16197 | -3.539 | -0.6046 | Yes | ||
| 44 | NT5C3 | 16356 | -3.765 | -0.6039 | Yes | ||
| 45 | POLR2A | 16496 | -3.999 | -0.6015 | Yes | ||
| 46 | POLR3A | 16519 | -4.032 | -0.5927 | Yes | ||
| 47 | POLR3K | 16580 | -4.126 | -0.5858 | Yes | ||
| 48 | POLE | 16901 | -4.758 | -0.5913 | Yes | ||
| 49 | POLR2G | 16995 | -4.926 | -0.5842 | Yes | ||
| 50 | NME4 | 17046 | -5.056 | -0.5744 | Yes | ||
| 51 | POLR1D | 17100 | -5.209 | -0.5644 | Yes | ||
| 52 | ENTPD4 | 17190 | -5.442 | -0.5558 | Yes | ||
| 53 | POLA2 | 17214 | -5.514 | -0.5434 | Yes | ||
| 54 | POLR2K | 17275 | -5.655 | -0.5327 | Yes | ||
| 55 | DTYMK | 17280 | -5.669 | -0.5190 | Yes | ||
| 56 | POLR2E | 17727 | -7.209 | -0.5252 | Yes | ||
| 57 | UCK1 | 17746 | -7.277 | -0.5083 | Yes | ||
| 58 | POLE3 | 17784 | -7.429 | -0.4919 | Yes | ||
| 59 | NT5C | 17830 | -7.654 | -0.4755 | Yes | ||
| 60 | RFC5 | 17863 | -7.807 | -0.4579 | Yes | ||
| 61 | NME7 | 17964 | -8.320 | -0.4428 | Yes | ||
| 62 | UMPS | 17983 | -8.435 | -0.4230 | Yes | ||
| 63 | POLR2J | 18064 | -8.947 | -0.4052 | Yes | ||
| 64 | NME6 | 18123 | -9.359 | -0.3853 | Yes | ||
| 65 | POLR2H | 18157 | -9.680 | -0.3632 | Yes | ||
| 66 | CAD | 18228 | -10.256 | -0.3416 | Yes | ||
| 67 | DHODH | 18247 | -10.417 | -0.3169 | Yes | ||
| 68 | POLD2 | 18305 | -10.925 | -0.2930 | Yes | ||
| 69 | POLR1A | 18417 | -12.389 | -0.2685 | Yes | ||
| 70 | NME2 | 18459 | -13.123 | -0.2383 | Yes | ||
| 71 | NME1 | 18468 | -13.304 | -0.2059 | Yes | ||
| 72 | POLR3H | 18494 | -13.702 | -0.1735 | Yes | ||
| 73 | DCTD | 18527 | -15.032 | -0.1381 | Yes | ||
| 74 | POLR3B | 18539 | -15.622 | -0.1002 | Yes | ||
| 75 | POLR1B | 18596 | -21.115 | -0.0511 | Yes | ||
| 76 | CTPS | 18597 | -21.144 | 0.0010 | Yes |
