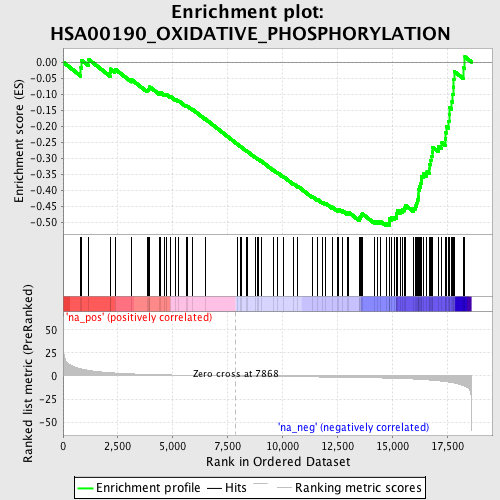
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA00190_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.5119837 |
| Normalized Enrichment Score (NES) | -1.7813028 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.020131625 |
| FWER p-Value | 0.36 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COX6B2 | 811 | 7.682 | -0.0180 | No | ||
| 2 | TCIRG1 | 839 | 7.526 | 0.0057 | No | ||
| 3 | COX7A1 | 1172 | 6.032 | 0.0080 | No | ||
| 4 | ATP6V0D1 | 2150 | 3.549 | -0.0328 | No | ||
| 5 | ATP6V1G2 | 2160 | 3.535 | -0.0215 | No | ||
| 6 | NDUFS5 | 2364 | 3.191 | -0.0217 | No | ||
| 7 | NDUFA13 | 3115 | 2.288 | -0.0546 | No | ||
| 8 | ATP6V0A2 | 3829 | 1.691 | -0.0874 | No | ||
| 9 | ATP6V1D | 3882 | 1.650 | -0.0847 | No | ||
| 10 | ATP6V0C | 3920 | 1.626 | -0.0812 | No | ||
| 11 | UQCRFS1 | 3953 | 1.608 | -0.0776 | No | ||
| 12 | ATP12A | 4385 | 1.357 | -0.0963 | No | ||
| 13 | SDHA | 4451 | 1.310 | -0.0954 | No | ||
| 14 | UQCRH | 4640 | 1.202 | -0.1015 | No | ||
| 15 | ATP6V1B2 | 4692 | 1.181 | -0.1003 | No | ||
| 16 | ATP4A | 4905 | 1.082 | -0.1081 | No | ||
| 17 | UQCRB | 5134 | 0.977 | -0.1172 | No | ||
| 18 | COX8C | 5270 | 0.911 | -0.1214 | No | ||
| 19 | ATP6V1H | 5605 | 0.772 | -0.1369 | No | ||
| 20 | COX8A | 5662 | 0.749 | -0.1374 | No | ||
| 21 | NDUFS4 | 5894 | 0.667 | -0.1476 | No | ||
| 22 | NDUFA6 | 6467 | 0.445 | -0.1770 | No | ||
| 23 | ATP6V1E2 | 7951 | -0.024 | -0.2570 | No | ||
| 24 | ATP6V1G3 | 8103 | -0.076 | -0.2649 | No | ||
| 25 | ATP4B | 8140 | -0.085 | -0.2666 | No | ||
| 26 | NDUFA4 | 8362 | -0.152 | -0.2780 | No | ||
| 27 | ATP6V1E1 | 8400 | -0.162 | -0.2795 | No | ||
| 28 | ATP6V1B1 | 8757 | -0.273 | -0.2978 | No | ||
| 29 | NDUFA3 | 8853 | -0.301 | -0.3019 | No | ||
| 30 | NDUFS1 | 8907 | -0.316 | -0.3037 | No | ||
| 31 | NDUFA1 | 9024 | -0.348 | -0.3088 | No | ||
| 32 | NDUFB11 | 9578 | -0.509 | -0.3370 | No | ||
| 33 | ATP6AP1 | 9789 | -0.570 | -0.3464 | No | ||
| 34 | COX4I2 | 10059 | -0.646 | -0.3588 | No | ||
| 35 | ATP6V0A4 | 10515 | -0.771 | -0.3807 | No | ||
| 36 | ATP6V1C2 | 10702 | -0.824 | -0.3880 | No | ||
| 37 | ATP6V0D2 | 11357 | -1.007 | -0.4200 | No | ||
| 38 | NDUFS8 | 11611 | -1.076 | -0.4300 | No | ||
| 39 | COX4I1 | 11838 | -1.149 | -0.4384 | No | ||
| 40 | ATP6V1A | 11957 | -1.185 | -0.4408 | No | ||
| 41 | ATP5C1 | 12257 | -1.274 | -0.4527 | No | ||
| 42 | ATP5H | 12498 | -1.348 | -0.4611 | No | ||
| 43 | ATP5B | 12557 | -1.363 | -0.4597 | No | ||
| 44 | NDUFB7 | 12734 | -1.422 | -0.4644 | No | ||
| 45 | NDUFA2 | 12942 | -1.490 | -0.4706 | No | ||
| 46 | ATP6V1G1 | 13026 | -1.516 | -0.4700 | No | ||
| 47 | NDUFA11 | 13513 | -1.697 | -0.4905 | No | ||
| 48 | ATP5G2 | 13518 | -1.699 | -0.4851 | No | ||
| 49 | UQCRC1 | 13571 | -1.722 | -0.4821 | No | ||
| 50 | SDHC | 13588 | -1.726 | -0.4772 | No | ||
| 51 | NDUFB10 | 13634 | -1.741 | -0.4738 | No | ||
| 52 | NDUFB5 | 14186 | -1.981 | -0.4969 | No | ||
| 53 | ATP6V1F | 14341 | -2.061 | -0.4983 | No | ||
| 54 | COX5B | 14480 | -2.133 | -0.4986 | No | ||
| 55 | COX6C | 14729 | -2.267 | -0.5044 | Yes | ||
| 56 | NDUFC2 | 14860 | -2.343 | -0.5035 | Yes | ||
| 57 | NDUFC1 | 14878 | -2.353 | -0.4966 | Yes | ||
| 58 | COX7C | 14883 | -2.355 | -0.4889 | Yes | ||
| 59 | NDUFB2 | 14977 | -2.412 | -0.4858 | Yes | ||
| 60 | ATP5J | 15114 | -2.506 | -0.4848 | Yes | ||
| 61 | ATP6V0A1 | 15191 | -2.563 | -0.4803 | Yes | ||
| 62 | NDUFB8 | 15203 | -2.568 | -0.4723 | Yes | ||
| 63 | ATP5E | 15220 | -2.582 | -0.4645 | Yes | ||
| 64 | NDUFB9 | 15361 | -2.690 | -0.4630 | Yes | ||
| 65 | ATP5J2 | 15475 | -2.770 | -0.4598 | Yes | ||
| 66 | ATP5A1 | 15576 | -2.855 | -0.4557 | Yes | ||
| 67 | NDUFA5 | 15617 | -2.892 | -0.4481 | Yes | ||
| 68 | NDUFA7 | 15986 | -3.280 | -0.4570 | Yes | ||
| 69 | ATP5O | 16055 | -3.362 | -0.4494 | Yes | ||
| 70 | ATP5G3 | 16106 | -3.421 | -0.4406 | Yes | ||
| 71 | ATP5D | 16132 | -3.445 | -0.4304 | Yes | ||
| 72 | ATP5I | 16179 | -3.515 | -0.4211 | Yes | ||
| 73 | ATP6V0E1 | 16198 | -3.542 | -0.4102 | Yes | ||
| 74 | ATP6V0B | 16200 | -3.544 | -0.3984 | Yes | ||
| 75 | COX6A2 | 16234 | -3.593 | -0.3881 | Yes | ||
| 76 | COX7B | 16303 | -3.686 | -0.3794 | Yes | ||
| 77 | ATP5F1 | 16323 | -3.717 | -0.3680 | Yes | ||
| 78 | COX5A | 16336 | -3.739 | -0.3561 | Yes | ||
| 79 | COX6A1 | 16441 | -3.899 | -0.3487 | Yes | ||
| 80 | NDUFS7 | 16583 | -4.134 | -0.3424 | Yes | ||
| 81 | NDUFB4 | 16689 | -4.339 | -0.3335 | Yes | ||
| 82 | COX15 | 16694 | -4.351 | -0.3192 | Yes | ||
| 83 | COX6B1 | 16739 | -4.432 | -0.3067 | Yes | ||
| 84 | NDUFA10 | 16776 | -4.522 | -0.2935 | Yes | ||
| 85 | SDHD | 16828 | -4.632 | -0.2807 | Yes | ||
| 86 | NDUFAB1 | 16844 | -4.660 | -0.2659 | Yes | ||
| 87 | COX10 | 17104 | -5.217 | -0.2624 | Yes | ||
| 88 | NDUFS2 | 17262 | -5.625 | -0.2520 | Yes | ||
| 89 | COX7A2 | 17413 | -6.040 | -0.2398 | Yes | ||
| 90 | ATP6V1C1 | 17439 | -6.111 | -0.2207 | Yes | ||
| 91 | UQCR | 17460 | -6.186 | -0.2010 | Yes | ||
| 92 | NDUFA8 | 17558 | -6.546 | -0.1843 | Yes | ||
| 93 | NDUFV1 | 17595 | -6.695 | -0.1638 | Yes | ||
| 94 | NDUFB3 | 17605 | -6.724 | -0.1418 | Yes | ||
| 95 | UQCRC2 | 17720 | -7.160 | -0.1239 | Yes | ||
| 96 | ATP5G1 | 17758 | -7.336 | -0.1013 | Yes | ||
| 97 | COX17 | 17785 | -7.435 | -0.0778 | Yes | ||
| 98 | NDUFA9 | 17795 | -7.504 | -0.0531 | Yes | ||
| 99 | CYC1 | 17821 | -7.633 | -0.0289 | Yes | ||
| 100 | SDHB | 18233 | -10.325 | -0.0164 | Yes | ||
| 101 | NDUFS3 | 18313 | -11.057 | 0.0164 | Yes |
