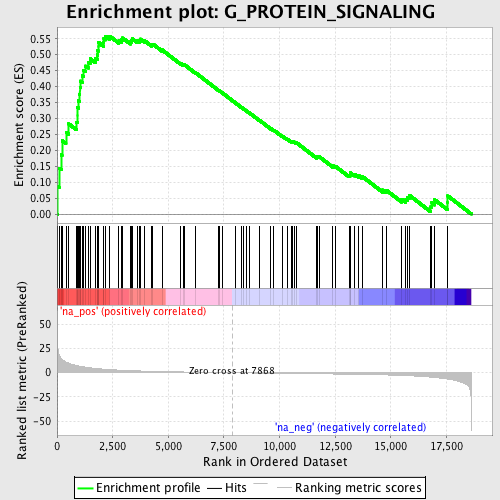
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | G_PROTEIN_SIGNALING |
| Enrichment Score (ES) | 0.55817485 |
| Normalized Enrichment Score (NES) | 1.8072768 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.044047758 |
| FWER p-Value | 0.398 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GNA15 | 26 | 26.161 | 0.0862 | Yes | ||
| 2 | PRKCH | 92 | 18.399 | 0.1443 | Yes | ||
| 3 | PRKCQ | 196 | 14.046 | 0.1858 | Yes | ||
| 4 | IL18BP | 223 | 13.597 | 0.2299 | Yes | ||
| 5 | PRKACB | 400 | 10.923 | 0.2570 | Yes | ||
| 6 | PDE4D | 524 | 9.718 | 0.2829 | Yes | ||
| 7 | RRAS | 873 | 7.329 | 0.2887 | Yes | ||
| 8 | GNAQ | 918 | 7.135 | 0.3102 | Yes | ||
| 9 | PDE4A | 919 | 7.132 | 0.3341 | Yes | ||
| 10 | SLC9A1 | 957 | 6.924 | 0.3552 | Yes | ||
| 11 | GNB2 | 1001 | 6.736 | 0.3755 | Yes | ||
| 12 | PRKCB1 | 1028 | 6.604 | 0.3962 | Yes | ||
| 13 | ARHGEF1 | 1043 | 6.559 | 0.4174 | Yes | ||
| 14 | PRKAR1A | 1129 | 6.235 | 0.4337 | Yes | ||
| 15 | ITPR1 | 1193 | 5.947 | 0.4502 | Yes | ||
| 16 | PRKCD | 1279 | 5.615 | 0.4644 | Yes | ||
| 17 | CALM3 | 1400 | 5.225 | 0.4755 | Yes | ||
| 18 | ADCY6 | 1486 | 4.989 | 0.4876 | Yes | ||
| 19 | PLCB3 | 1732 | 4.377 | 0.4890 | Yes | ||
| 20 | PDE4B | 1805 | 4.224 | 0.4993 | Yes | ||
| 21 | ADCY9 | 1827 | 4.185 | 0.5122 | Yes | ||
| 22 | KRAS | 1847 | 4.132 | 0.5250 | Yes | ||
| 23 | PPP3CC | 1849 | 4.130 | 0.5387 | Yes | ||
| 24 | AKAP12 | 2090 | 3.653 | 0.5380 | Yes | ||
| 25 | AKAP9 | 2101 | 3.640 | 0.5497 | Yes | ||
| 26 | GNB5 | 2171 | 3.516 | 0.5577 | Yes | ||
| 27 | GNG12 | 2362 | 3.194 | 0.5582 | Yes | ||
| 28 | PPP3CA | 2779 | 2.647 | 0.5446 | No | ||
| 29 | GNA13 | 2899 | 2.500 | 0.5465 | No | ||
| 30 | PDE1B | 2943 | 2.454 | 0.5524 | No | ||
| 31 | GNA12 | 3300 | 2.106 | 0.5403 | No | ||
| 32 | GNG10 | 3349 | 2.065 | 0.5446 | No | ||
| 33 | GNG5 | 3379 | 2.034 | 0.5499 | No | ||
| 34 | CALM2 | 3612 | 1.847 | 0.5435 | No | ||
| 35 | PRKD3 | 3698 | 1.783 | 0.5449 | No | ||
| 36 | KCNJ3 | 3730 | 1.762 | 0.5491 | No | ||
| 37 | GNB1 | 3917 | 1.627 | 0.5446 | No | ||
| 38 | GNA14 | 4251 | 1.435 | 0.5314 | No | ||
| 39 | PDE4C | 4306 | 1.404 | 0.5332 | No | ||
| 40 | AKAP3 | 4715 | 1.171 | 0.5151 | No | ||
| 41 | AKAP4 | 5566 | 0.786 | 0.4719 | No | ||
| 42 | ADCY5 | 5691 | 0.736 | 0.4676 | No | ||
| 43 | GNG4 | 5706 | 0.727 | 0.4693 | No | ||
| 44 | PRKD1 | 6234 | 0.529 | 0.4427 | No | ||
| 45 | AKAP10 | 7240 | 0.185 | 0.3890 | No | ||
| 46 | GNAI3 | 7291 | 0.172 | 0.3869 | No | ||
| 47 | GNG3 | 7415 | 0.137 | 0.3807 | No | ||
| 48 | PDE8A | 8028 | -0.053 | 0.3479 | No | ||
| 49 | PRKCI | 8291 | -0.129 | 0.3342 | No | ||
| 50 | PDE1C | 8376 | -0.156 | 0.3302 | No | ||
| 51 | PRKCA | 8501 | -0.195 | 0.3242 | No | ||
| 52 | ADCY8 | 8632 | -0.234 | 0.3179 | No | ||
| 53 | GNAO1 | 9081 | -0.365 | 0.2950 | No | ||
| 54 | GNG7 | 9610 | -0.518 | 0.2682 | No | ||
| 55 | GNGT1 | 9733 | -0.553 | 0.2635 | No | ||
| 56 | PDE7B | 10133 | -0.665 | 0.2442 | No | ||
| 57 | PDE1A | 10345 | -0.723 | 0.2352 | No | ||
| 58 | PRKACA | 10529 | -0.773 | 0.2279 | No | ||
| 59 | PRKAR1B | 10560 | -0.783 | 0.2289 | No | ||
| 60 | PRKCZ | 10666 | -0.815 | 0.2260 | No | ||
| 61 | RHOA | 10740 | -0.836 | 0.2249 | No | ||
| 62 | PRKCE | 11664 | -1.095 | 0.1787 | No | ||
| 63 | AKAP11 | 11701 | -1.105 | 0.1805 | No | ||
| 64 | GNB3 | 11788 | -1.135 | 0.1796 | No | ||
| 65 | GNA11 | 12396 | -1.317 | 0.1513 | No | ||
| 66 | CALM1 | 12515 | -1.352 | 0.1495 | No | ||
| 67 | PDE8B | 13130 | -1.549 | 0.1215 | No | ||
| 68 | GNAZ | 13172 | -1.565 | 0.1246 | No | ||
| 69 | ADCY4 | 13175 | -1.566 | 0.1297 | No | ||
| 70 | AKAP8 | 13369 | -1.641 | 0.1248 | No | ||
| 71 | AKAP7 | 13540 | -1.709 | 0.1213 | No | ||
| 72 | GNGT2 | 13708 | -1.773 | 0.1183 | No | ||
| 73 | HRAS | 14637 | -2.213 | 0.0756 | No | ||
| 74 | ADCY2 | 14786 | -2.303 | 0.0753 | No | ||
| 75 | ADCY3 | 15489 | -2.784 | 0.0468 | No | ||
| 76 | GNG13 | 15674 | -2.936 | 0.0467 | No | ||
| 77 | ADCY7 | 15754 | -3.017 | 0.0525 | No | ||
| 78 | USP5 | 15824 | -3.116 | 0.0592 | No | ||
| 79 | NRAS | 16794 | -4.560 | 0.0222 | No | ||
| 80 | PDE7A | 16842 | -4.656 | 0.0353 | No | ||
| 81 | PRKAR2B | 16973 | -4.887 | 0.0446 | No | ||
| 82 | PRKAR2A | 17556 | -6.541 | 0.0351 | No | ||
| 83 | AKAP1 | 17563 | -6.577 | 0.0568 | No |
