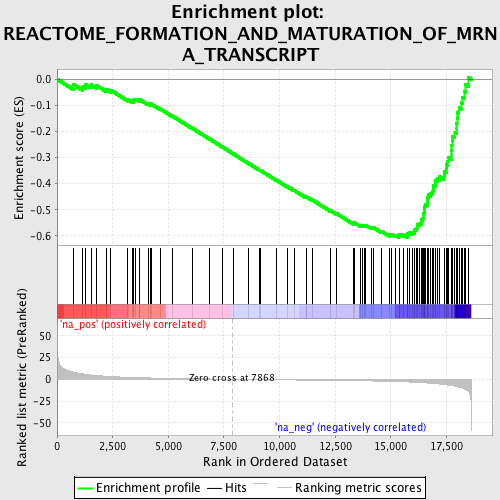
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | -0.60597473 |
| Normalized Enrichment Score (NES) | -2.1095722 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ERCC2 | 737 | 8.125 | -0.0197 | No | ||
| 2 | CD2BP2 | 1161 | 6.083 | -0.0275 | No | ||
| 3 | TCEB2 | 1286 | 5.588 | -0.0203 | No | ||
| 4 | RBM5 | 1530 | 4.862 | -0.0214 | No | ||
| 5 | CCNT1 | 1773 | 4.279 | -0.0239 | No | ||
| 6 | ELL | 2233 | 3.443 | -0.0401 | No | ||
| 7 | PCBP2 | 2410 | 3.124 | -0.0419 | No | ||
| 8 | SMC1A | 3180 | 2.219 | -0.0780 | No | ||
| 9 | PCF11 | 3408 | 2.006 | -0.0852 | No | ||
| 10 | TCEB3 | 3451 | 1.968 | -0.0826 | No | ||
| 11 | RBMX | 3452 | 1.968 | -0.0778 | No | ||
| 12 | PABPN1 | 3509 | 1.927 | -0.0760 | No | ||
| 13 | PAPOLA | 3689 | 1.786 | -0.0813 | No | ||
| 14 | CCAR1 | 3706 | 1.779 | -0.0777 | No | ||
| 15 | SRRM1 | 4088 | 1.531 | -0.0945 | No | ||
| 16 | CCNT2 | 4208 | 1.462 | -0.0973 | No | ||
| 17 | SF3B2 | 4220 | 1.454 | -0.0943 | No | ||
| 18 | SFRS7 | 4635 | 1.206 | -0.1137 | No | ||
| 19 | SF4 | 5198 | 0.946 | -0.1417 | No | ||
| 20 | POLR2C | 6074 | 0.590 | -0.1875 | No | ||
| 21 | FUS | 6864 | 0.309 | -0.2293 | No | ||
| 22 | CSTF2 | 7452 | 0.125 | -0.2607 | No | ||
| 23 | SNRPB2 | 7945 | -0.021 | -0.2873 | No | ||
| 24 | TAF13 | 8618 | -0.227 | -0.3230 | No | ||
| 25 | SFRS1 | 9103 | -0.371 | -0.3482 | No | ||
| 26 | TAF4 | 9141 | -0.382 | -0.3493 | No | ||
| 27 | GTF2B | 9878 | -0.595 | -0.3875 | No | ||
| 28 | PTBP1 | 10352 | -0.724 | -0.4113 | No | ||
| 29 | MNAT1 | 10657 | -0.812 | -0.4257 | No | ||
| 30 | SF3B4 | 11197 | -0.956 | -0.4525 | No | ||
| 31 | SF3B1 | 11205 | -0.960 | -0.4505 | No | ||
| 32 | SNRPE | 11463 | -1.035 | -0.4618 | No | ||
| 33 | POLR2B | 12304 | -1.288 | -0.5040 | No | ||
| 34 | DNAJC8 | 12537 | -1.357 | -0.5131 | No | ||
| 35 | SFRS9 | 13314 | -1.619 | -0.5510 | No | ||
| 36 | GTF2F1 | 13366 | -1.640 | -0.5497 | No | ||
| 37 | GTF2A2 | 13633 | -1.740 | -0.5598 | No | ||
| 38 | SFRS5 | 13713 | -1.776 | -0.5596 | No | ||
| 39 | POLR2I | 13802 | -1.813 | -0.5599 | No | ||
| 40 | CDK7 | 13862 | -1.838 | -0.5585 | No | ||
| 41 | PCBP1 | 14116 | -1.952 | -0.5674 | No | ||
| 42 | PRPF8 | 14202 | -1.989 | -0.5670 | No | ||
| 43 | NUDT21 | 14593 | -2.191 | -0.5827 | No | ||
| 44 | SFRS3 | 14934 | -2.388 | -0.5951 | No | ||
| 45 | DHX9 | 15026 | -2.445 | -0.5940 | No | ||
| 46 | U2AF2 | 15211 | -2.574 | -0.5975 | No | ||
| 47 | TAF10 | 15368 | -2.693 | -0.5993 | Yes | ||
| 48 | SUPT4H1 | 15386 | -2.708 | -0.5935 | Yes | ||
| 49 | THOC4 | 15573 | -2.853 | -0.5965 | Yes | ||
| 50 | SF3A3 | 15730 | -2.996 | -0.5975 | Yes | ||
| 51 | CPSF1 | 15753 | -3.016 | -0.5912 | Yes | ||
| 52 | GTF2A1 | 15820 | -3.112 | -0.5871 | Yes | ||
| 53 | TAF9 | 15983 | -3.278 | -0.5877 | Yes | ||
| 54 | TAF6 | 16053 | -3.360 | -0.5831 | Yes | ||
| 55 | SNRPF | 16058 | -3.366 | -0.5750 | Yes | ||
| 56 | GTF2H1 | 16160 | -3.494 | -0.5718 | Yes | ||
| 57 | RNMT | 16181 | -3.518 | -0.5642 | Yes | ||
| 58 | SNRPD3 | 16192 | -3.532 | -0.5560 | Yes | ||
| 59 | RNPS1 | 16284 | -3.659 | -0.5518 | Yes | ||
| 60 | CSTF1 | 16381 | -3.802 | -0.5476 | Yes | ||
| 61 | SF3A1 | 16384 | -3.810 | -0.5383 | Yes | ||
| 62 | SF3A2 | 16439 | -3.897 | -0.5315 | Yes | ||
| 63 | SNRPG | 16459 | -3.937 | -0.5228 | Yes | ||
| 64 | NFX1 | 16462 | -3.940 | -0.5132 | Yes | ||
| 65 | POLR2A | 16496 | -3.999 | -0.5050 | Yes | ||
| 66 | CCNH | 16501 | -4.006 | -0.4953 | Yes | ||
| 67 | TCEB1 | 16512 | -4.017 | -0.4859 | Yes | ||
| 68 | CDK9 | 16560 | -4.094 | -0.4783 | Yes | ||
| 69 | RDBP | 16641 | -4.232 | -0.4722 | Yes | ||
| 70 | SNRPA | 16650 | -4.254 | -0.4621 | Yes | ||
| 71 | METTL3 | 16659 | -4.273 | -0.4519 | Yes | ||
| 72 | MAGOH | 16695 | -4.351 | -0.4430 | Yes | ||
| 73 | SFRS6 | 16781 | -4.534 | -0.4364 | Yes | ||
| 74 | TAF11 | 16875 | -4.711 | -0.4297 | Yes | ||
| 75 | TAF5 | 16902 | -4.758 | -0.4194 | Yes | ||
| 76 | CSTF3 | 16921 | -4.791 | -0.4085 | Yes | ||
| 77 | POLR2G | 16995 | -4.926 | -0.4002 | Yes | ||
| 78 | GTF2E2 | 17001 | -4.949 | -0.3882 | Yes | ||
| 79 | GTF2E1 | 17083 | -5.149 | -0.3799 | Yes | ||
| 80 | RNGTT | 17202 | -5.477 | -0.3727 | Yes | ||
| 81 | SF3B3 | 17391 | -5.985 | -0.3680 | Yes | ||
| 82 | NCBP2 | 17401 | -6.011 | -0.3536 | Yes | ||
| 83 | TBP | 17498 | -6.354 | -0.3431 | Yes | ||
| 84 | SNRPD2 | 17510 | -6.413 | -0.3278 | Yes | ||
| 85 | ERCC3 | 17559 | -6.558 | -0.3141 | Yes | ||
| 86 | SNRPA1 | 17572 | -6.605 | -0.2984 | Yes | ||
| 87 | POLR2E | 17727 | -7.209 | -0.2889 | Yes | ||
| 88 | YBX1 | 17728 | -7.210 | -0.2710 | Yes | ||
| 89 | TH1L | 17745 | -7.277 | -0.2539 | Yes | ||
| 90 | SFRS2 | 17755 | -7.315 | -0.2362 | Yes | ||
| 91 | PHF5A | 17762 | -7.350 | -0.2184 | Yes | ||
| 92 | U2AF1 | 17883 | -7.920 | -0.2052 | Yes | ||
| 93 | PRPF4 | 17956 | -8.274 | -0.1886 | Yes | ||
| 94 | GTF2H3 | 17958 | -8.285 | -0.1682 | Yes | ||
| 95 | TXNL4A | 17976 | -8.391 | -0.1483 | Yes | ||
| 96 | TAF12 | 17992 | -8.494 | -0.1281 | Yes | ||
| 97 | POLR2J | 18064 | -8.947 | -0.1098 | Yes | ||
| 98 | POLR2H | 18157 | -9.680 | -0.0908 | Yes | ||
| 99 | SNRPB | 18222 | -10.194 | -0.0690 | Yes | ||
| 100 | EFTUD2 | 18300 | -10.862 | -0.0462 | Yes | ||
| 101 | GTF2H2 | 18357 | -11.616 | -0.0205 | Yes | ||
| 102 | GTF2F2 | 18499 | -13.900 | 0.0063 | Yes |
