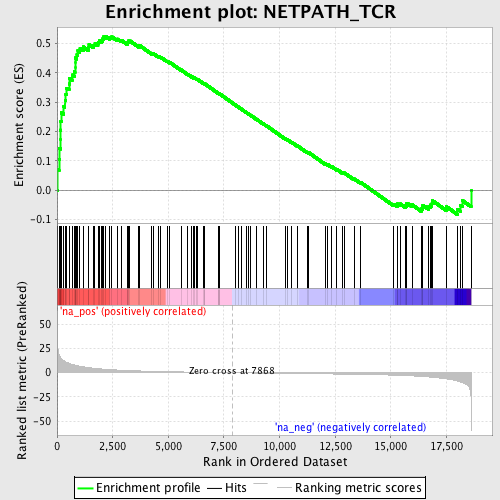
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | 0.525252 |
| Normalized Enrichment Score (NES) | 1.8120743 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.052525457 |
| FWER p-Value | 0.516 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0704 | Yes | ||
| 2 | PTPN22 | 89 | 18.514 | 0.1054 | Yes | ||
| 3 | JAK3 | 108 | 17.689 | 0.1419 | Yes | ||
| 4 | PSTPIP1 | 145 | 15.490 | 0.1728 | Yes | ||
| 5 | CISH | 151 | 15.397 | 0.2051 | Yes | ||
| 6 | NEDD9 | 165 | 14.716 | 0.2356 | Yes | ||
| 7 | PRKCQ | 196 | 14.046 | 0.2637 | Yes | ||
| 8 | GIT2 | 300 | 12.327 | 0.2843 | Yes | ||
| 9 | VAV1 | 354 | 11.615 | 0.3060 | Yes | ||
| 10 | CD5 | 391 | 11.048 | 0.3275 | Yes | ||
| 11 | LAT | 432 | 10.644 | 0.3479 | Yes | ||
| 12 | DEF6 | 540 | 9.514 | 0.3622 | Yes | ||
| 13 | FYN | 553 | 9.371 | 0.3815 | Yes | ||
| 14 | SH3BP2 | 676 | 8.459 | 0.3928 | Yes | ||
| 15 | LCP2 | 783 | 7.814 | 0.4036 | Yes | ||
| 16 | STAT5A | 807 | 7.710 | 0.4187 | Yes | ||
| 17 | PTPN12 | 810 | 7.695 | 0.4349 | Yes | ||
| 18 | SH2D2A | 813 | 7.647 | 0.4510 | Yes | ||
| 19 | WASF2 | 891 | 7.226 | 0.4621 | Yes | ||
| 20 | WAS | 934 | 7.059 | 0.4748 | Yes | ||
| 21 | ZAP70 | 1027 | 6.606 | 0.4838 | Yes | ||
| 22 | LCK | 1176 | 6.008 | 0.4886 | Yes | ||
| 23 | HOMER3 | 1408 | 5.212 | 0.4871 | Yes | ||
| 24 | RASA1 | 1431 | 5.144 | 0.4968 | Yes | ||
| 25 | PTK2B | 1651 | 4.557 | 0.4947 | Yes | ||
| 26 | CREBBP | 1702 | 4.447 | 0.5014 | Yes | ||
| 27 | SHB | 1838 | 4.155 | 0.5029 | Yes | ||
| 28 | PAG1 | 1882 | 4.067 | 0.5092 | Yes | ||
| 29 | ABL1 | 2004 | 3.811 | 0.5107 | Yes | ||
| 30 | SH2D3C | 2058 | 3.709 | 0.5157 | Yes | ||
| 31 | SOS2 | 2078 | 3.676 | 0.5225 | Yes | ||
| 32 | GRB2 | 2166 | 3.530 | 0.5253 | Yes | ||
| 33 | ABI1 | 2360 | 3.195 | 0.5216 | No | ||
| 34 | EVL | 2463 | 3.060 | 0.5226 | No | ||
| 35 | DNM2 | 2703 | 2.729 | 0.5154 | No | ||
| 36 | CRKL | 2883 | 2.519 | 0.5111 | No | ||
| 37 | CDC42 | 3158 | 2.237 | 0.5010 | No | ||
| 38 | CREB1 | 3178 | 2.219 | 0.5047 | No | ||
| 39 | DOCK2 | 3190 | 2.206 | 0.5088 | No | ||
| 40 | JUN | 3275 | 2.124 | 0.5088 | No | ||
| 41 | GRAP2 | 3668 | 1.800 | 0.4914 | No | ||
| 42 | CD247 | 3691 | 1.785 | 0.4940 | No | ||
| 43 | PIK3R1 | 4260 | 1.432 | 0.4663 | No | ||
| 44 | GAB2 | 4314 | 1.401 | 0.4664 | No | ||
| 45 | FYB | 4551 | 1.249 | 0.4563 | No | ||
| 46 | SOS1 | 4644 | 1.201 | 0.4539 | No | ||
| 47 | CD3G | 4946 | 1.062 | 0.4399 | No | ||
| 48 | MAP2K1 | 5069 | 1.006 | 0.4354 | No | ||
| 49 | INSL3 | 5570 | 0.784 | 0.4101 | No | ||
| 50 | PTPN3 | 5878 | 0.672 | 0.3949 | No | ||
| 51 | PTPRC | 6059 | 0.599 | 0.3865 | No | ||
| 52 | ARHGDIB | 6150 | 0.562 | 0.3828 | No | ||
| 53 | FOS | 6176 | 0.553 | 0.3826 | No | ||
| 54 | TXK | 6276 | 0.516 | 0.3784 | No | ||
| 55 | RAPGEF1 | 6316 | 0.498 | 0.3773 | No | ||
| 56 | CD3D | 6580 | 0.407 | 0.3640 | No | ||
| 57 | WIPF1 | 6627 | 0.391 | 0.3623 | No | ||
| 58 | LYN | 6633 | 0.389 | 0.3629 | No | ||
| 59 | PLCG1 | 7237 | 0.186 | 0.3307 | No | ||
| 60 | UNC119 | 7249 | 0.183 | 0.3305 | No | ||
| 61 | TUBB2A | 7289 | 0.173 | 0.3288 | No | ||
| 62 | BCL10 | 7310 | 0.166 | 0.3280 | No | ||
| 63 | SKAP2 | 8006 | -0.044 | 0.2906 | No | ||
| 64 | ITK | 8160 | -0.091 | 0.2825 | No | ||
| 65 | SLA | 8271 | -0.125 | 0.2768 | No | ||
| 66 | ARHGEF7 | 8504 | -0.195 | 0.2647 | No | ||
| 67 | STAT5B | 8605 | -0.224 | 0.2598 | No | ||
| 68 | PTPRH | 8704 | -0.258 | 0.2550 | No | ||
| 69 | CD3E | 8947 | -0.329 | 0.2427 | No | ||
| 70 | CD2AP | 9269 | -0.421 | 0.2262 | No | ||
| 71 | CEBPB | 9400 | -0.457 | 0.2201 | No | ||
| 72 | GRAP | 10268 | -0.699 | 0.1748 | No | ||
| 73 | STK39 | 10363 | -0.726 | 0.1713 | No | ||
| 74 | NCK1 | 10552 | -0.782 | 0.1628 | No | ||
| 75 | RAP1A | 10791 | -0.848 | 0.1517 | No | ||
| 76 | NFATC2 | 11235 | -0.968 | 0.1298 | No | ||
| 77 | PTPRJ | 11318 | -0.994 | 0.1275 | No | ||
| 78 | CD4 | 12066 | -1.218 | 0.0897 | No | ||
| 79 | SRC | 12150 | -1.245 | 0.0879 | No | ||
| 80 | CBL | 12327 | -1.294 | 0.0811 | No | ||
| 81 | ACP1 | 12574 | -1.367 | 0.0707 | No | ||
| 82 | SYK | 12848 | -1.460 | 0.0591 | No | ||
| 83 | SHC1 | 12913 | -1.483 | 0.0587 | No | ||
| 84 | DLG1 | 13358 | -1.638 | 0.0382 | No | ||
| 85 | AKT1 | 13635 | -1.741 | 0.0270 | No | ||
| 86 | ENAH | 15129 | -2.515 | -0.0483 | No | ||
| 87 | NCL | 15287 | -2.635 | -0.0512 | No | ||
| 88 | PIK3R2 | 15311 | -2.649 | -0.0468 | No | ||
| 89 | KHDRBS1 | 15419 | -2.733 | -0.0468 | No | ||
| 90 | TUBA4A | 15653 | -2.917 | -0.0532 | No | ||
| 91 | YWHAQ | 15717 | -2.978 | -0.0503 | No | ||
| 92 | DUSP3 | 15718 | -2.980 | -0.0440 | No | ||
| 93 | VASP | 15953 | -3.246 | -0.0498 | No | ||
| 94 | DBNL | 16363 | -3.776 | -0.0639 | No | ||
| 95 | RIPK2 | 16429 | -3.880 | -0.0592 | No | ||
| 96 | CBLB | 16437 | -3.895 | -0.0513 | No | ||
| 97 | PTPN6 | 16706 | -4.373 | -0.0565 | No | ||
| 98 | MAPK1 | 16773 | -4.517 | -0.0505 | No | ||
| 99 | PTPN11 | 16849 | -4.668 | -0.0447 | No | ||
| 100 | CRK | 16873 | -4.709 | -0.0359 | No | ||
| 101 | ARHGEF6 | 17493 | -6.346 | -0.0559 | No | ||
| 102 | CARD11 | 17995 | -8.517 | -0.0650 | No | ||
| 103 | DTX1 | 18121 | -9.347 | -0.0519 | No | ||
| 104 | MAP4K1 | 18224 | -10.205 | -0.0358 | No | ||
| 105 | PTK2 | 18609 | -26.874 | 0.0004 | No |