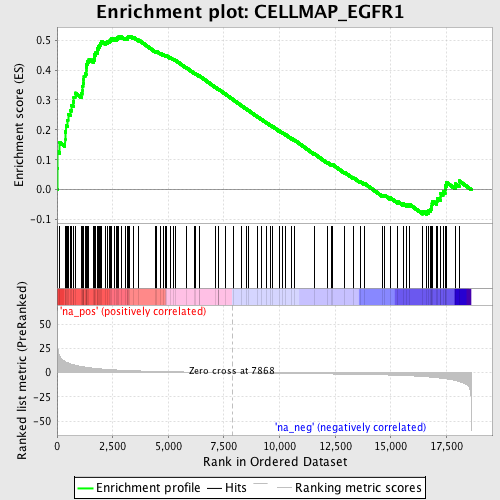
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CELLMAP_EGFR1 |
| Enrichment Score (ES) | 0.51361936 |
| Normalized Enrichment Score (NES) | 1.7958441 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.052831657 |
| FWER p-Value | 0.597 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 11 | 33.496 | 0.0701 | Yes | ||
| 2 | GAB1 | 24 | 27.320 | 0.1271 | Yes | ||
| 3 | GRB7 | 120 | 16.803 | 0.1574 | Yes | ||
| 4 | VAV1 | 354 | 11.615 | 0.1693 | Yes | ||
| 5 | STAT1 | 359 | 11.544 | 0.1935 | Yes | ||
| 6 | JAK2 | 399 | 10.951 | 0.2145 | Yes | ||
| 7 | SH3KBP1 | 465 | 10.267 | 0.2326 | Yes | ||
| 8 | STAT3 | 490 | 10.026 | 0.2525 | Yes | ||
| 9 | RALBP1 | 597 | 9.033 | 0.2658 | Yes | ||
| 10 | PIK3CG | 650 | 8.605 | 0.2812 | Yes | ||
| 11 | SMAD3 | 725 | 8.199 | 0.2945 | Yes | ||
| 12 | MAPK14 | 754 | 8.054 | 0.3100 | Yes | ||
| 13 | STAT5A | 807 | 7.710 | 0.3234 | Yes | ||
| 14 | TNIP1 | 1090 | 6.378 | 0.3217 | Yes | ||
| 15 | MAP3K14 | 1121 | 6.262 | 0.3332 | Yes | ||
| 16 | PRKAR1A | 1129 | 6.235 | 0.3460 | Yes | ||
| 17 | RPS6KA2 | 1164 | 6.074 | 0.3570 | Yes | ||
| 18 | SH3BGRL | 1197 | 5.931 | 0.3678 | Yes | ||
| 19 | SH3GL3 | 1207 | 5.894 | 0.3797 | Yes | ||
| 20 | RIPK1 | 1256 | 5.685 | 0.3891 | Yes | ||
| 21 | PLEC1 | 1305 | 5.506 | 0.3982 | Yes | ||
| 22 | STAT2 | 1309 | 5.493 | 0.4096 | Yes | ||
| 23 | WASL | 1327 | 5.444 | 0.4202 | Yes | ||
| 24 | MTA2 | 1370 | 5.306 | 0.4291 | Yes | ||
| 25 | RASA1 | 1431 | 5.144 | 0.4367 | Yes | ||
| 26 | PTK2B | 1651 | 4.557 | 0.4345 | Yes | ||
| 27 | NCK2 | 1677 | 4.496 | 0.4426 | Yes | ||
| 28 | MAPK3 | 1689 | 4.468 | 0.4515 | Yes | ||
| 29 | CAV2 | 1741 | 4.346 | 0.4579 | Yes | ||
| 30 | YWHAB | 1809 | 4.217 | 0.4632 | Yes | ||
| 31 | PAK1 | 1819 | 4.193 | 0.4715 | Yes | ||
| 32 | KRAS | 1847 | 4.132 | 0.4788 | Yes | ||
| 33 | EGF | 1914 | 3.994 | 0.4836 | Yes | ||
| 34 | ELK4 | 1954 | 3.914 | 0.4898 | Yes | ||
| 35 | MAP3K4 | 2000 | 3.822 | 0.4954 | Yes | ||
| 36 | GRB2 | 2166 | 3.530 | 0.4939 | Yes | ||
| 37 | JAK1 | 2268 | 3.366 | 0.4956 | Yes | ||
| 38 | ABI1 | 2360 | 3.195 | 0.4974 | Yes | ||
| 39 | RALGDS | 2418 | 3.116 | 0.5009 | Yes | ||
| 40 | PLD2 | 2452 | 3.076 | 0.5056 | Yes | ||
| 41 | SOCS3 | 2592 | 2.869 | 0.5042 | Yes | ||
| 42 | PLD1 | 2670 | 2.763 | 0.5058 | Yes | ||
| 43 | EEF1A1 | 2722 | 2.703 | 0.5088 | Yes | ||
| 44 | PLSCR1 | 2778 | 2.648 | 0.5114 | Yes | ||
| 45 | CRKL | 2883 | 2.519 | 0.5111 | Yes | ||
| 46 | HIP1 | 3057 | 2.338 | 0.5067 | Yes | ||
| 47 | CDC42 | 3158 | 2.237 | 0.5060 | Yes | ||
| 48 | CREB1 | 3178 | 2.219 | 0.5096 | Yes | ||
| 49 | ATF1 | 3197 | 2.194 | 0.5133 | Yes | ||
| 50 | JUN | 3275 | 2.124 | 0.5136 | Yes | ||
| 51 | CSK | 3411 | 2.002 | 0.5105 | No | ||
| 52 | RPS6KA1 | 3662 | 1.805 | 0.5008 | No | ||
| 53 | GRB14 | 4423 | 1.337 | 0.4626 | No | ||
| 54 | SHOC2 | 4456 | 1.309 | 0.4636 | No | ||
| 55 | SOS1 | 4644 | 1.201 | 0.4560 | No | ||
| 56 | SP1 | 4770 | 1.146 | 0.4517 | No | ||
| 57 | PITPNA | 4880 | 1.094 | 0.4481 | No | ||
| 58 | HDAC1 | 4932 | 1.069 | 0.4476 | No | ||
| 59 | GJA1 | 5076 | 1.003 | 0.4420 | No | ||
| 60 | RAC1 | 5218 | 0.936 | 0.4364 | No | ||
| 61 | CAV1 | 5305 | 0.896 | 0.4336 | No | ||
| 62 | RAF1 | 5794 | 0.702 | 0.4087 | No | ||
| 63 | FOS | 6176 | 0.553 | 0.3893 | No | ||
| 64 | PRKD1 | 6234 | 0.529 | 0.3873 | No | ||
| 65 | ARAF | 6377 | 0.479 | 0.3807 | No | ||
| 66 | SMAD2 | 6395 | 0.473 | 0.3807 | No | ||
| 67 | SPRY2 | 7110 | 0.230 | 0.3426 | No | ||
| 68 | PLCG1 | 7237 | 0.186 | 0.3362 | No | ||
| 69 | AP2A1 | 7239 | 0.186 | 0.3365 | No | ||
| 70 | MAP3K2 | 7250 | 0.182 | 0.3364 | No | ||
| 71 | PIK3CA | 7547 | 0.100 | 0.3206 | No | ||
| 72 | PTK6 | 7923 | -0.015 | 0.3004 | No | ||
| 73 | PRKCI | 8291 | -0.129 | 0.2808 | No | ||
| 74 | PRKCA | 8501 | -0.195 | 0.2699 | No | ||
| 75 | STAT5B | 8605 | -0.224 | 0.2648 | No | ||
| 76 | KLF11 | 8994 | -0.342 | 0.2446 | No | ||
| 77 | SIN3A | 9176 | -0.392 | 0.2356 | No | ||
| 78 | CEBPB | 9400 | -0.457 | 0.2245 | No | ||
| 79 | CBLC | 9592 | -0.514 | 0.2153 | No | ||
| 80 | DNM1 | 9699 | -0.544 | 0.2107 | No | ||
| 81 | ELF3 | 9977 | -0.620 | 0.1970 | No | ||
| 82 | MCF2 | 10127 | -0.663 | 0.1904 | No | ||
| 83 | ARF4 | 10275 | -0.701 | 0.1839 | No | ||
| 84 | NCK1 | 10552 | -0.782 | 0.1706 | No | ||
| 85 | PRKCZ | 10666 | -0.815 | 0.1662 | No | ||
| 86 | EGFR | 11555 | -1.062 | 0.1205 | No | ||
| 87 | SRC | 12150 | -1.245 | 0.0910 | No | ||
| 88 | CBL | 12327 | -1.294 | 0.0842 | No | ||
| 89 | SNCA | 12388 | -1.315 | 0.0838 | No | ||
| 90 | SHC1 | 12913 | -1.483 | 0.0586 | No | ||
| 91 | MAPK7 | 13332 | -1.628 | 0.0394 | No | ||
| 92 | AKT1 | 13635 | -1.741 | 0.0268 | No | ||
| 93 | EPS15 | 13811 | -1.818 | 0.0211 | No | ||
| 94 | HRAS | 14637 | -2.213 | -0.0188 | No | ||
| 95 | RAB5A | 14736 | -2.269 | -0.0193 | No | ||
| 96 | MAP2K2 | 14962 | -2.404 | -0.0264 | No | ||
| 97 | PIK3R2 | 15311 | -2.649 | -0.0396 | No | ||
| 98 | MYC | 15548 | -2.832 | -0.0464 | No | ||
| 99 | MAPK8 | 15721 | -2.984 | -0.0494 | No | ||
| 100 | CASP9 | 15860 | -3.149 | -0.0502 | No | ||
| 101 | CBLB | 16437 | -3.895 | -0.0731 | No | ||
| 102 | HAT1 | 16618 | -4.195 | -0.0740 | No | ||
| 103 | PTPN6 | 16706 | -4.373 | -0.0695 | No | ||
| 104 | NRAS | 16794 | -4.560 | -0.0645 | No | ||
| 105 | SOCS1 | 16831 | -4.638 | -0.0567 | No | ||
| 106 | PTPN11 | 16849 | -4.668 | -0.0478 | No | ||
| 107 | CRK | 16873 | -4.709 | -0.0391 | No | ||
| 108 | INPPL1 | 17070 | -5.124 | -0.0389 | No | ||
| 109 | RGS16 | 17080 | -5.145 | -0.0285 | No | ||
| 110 | PIK3R3 | 17231 | -5.558 | -0.0249 | No | ||
| 111 | TNK2 | 17233 | -5.560 | -0.0132 | No | ||
| 112 | RBBP7 | 17347 | -5.853 | -0.0069 | No | ||
| 113 | PKN2 | 17440 | -6.120 | 0.0010 | No | ||
| 114 | RALB | 17451 | -6.168 | 0.0135 | No | ||
| 115 | SNRPD2 | 17510 | -6.413 | 0.0239 | No | ||
| 116 | PIK3CB | 17904 | -7.974 | 0.0195 | No | ||
| 117 | STXBP1 | 18070 | -8.993 | 0.0295 | No |