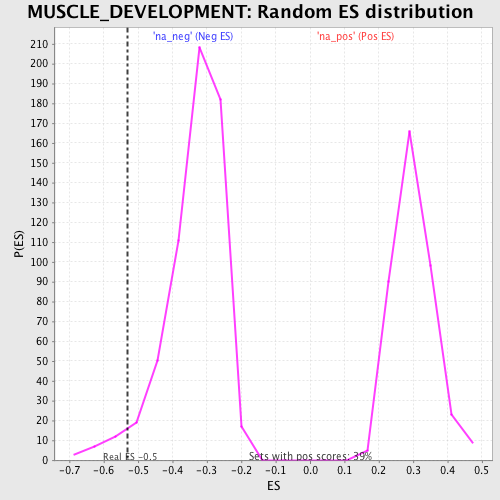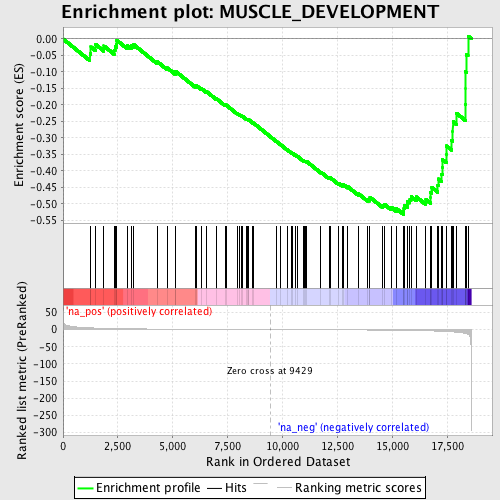
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MUSCLE_DEVELOPMENT |
| Enrichment Score (ES) | -0.53221697 |
| Normalized Enrichment Score (NES) | -1.5875323 |
| Nominal p-value | 0.036124796 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GAA | 1225 | 5.039 | -0.0437 | No | ||
| 2 | MAPK12 | 1269 | 4.916 | -0.0242 | No | ||
| 3 | MYEF2 | 1489 | 4.298 | -0.0169 | No | ||
| 4 | NRD1 | 1861 | 3.527 | -0.0213 | No | ||
| 5 | EGR3 | 2326 | 2.825 | -0.0338 | No | ||
| 6 | AEBP1 | 2369 | 2.767 | -0.0237 | No | ||
| 7 | IFRD1 | 2435 | 2.683 | -0.0153 | No | ||
| 8 | SVIL | 2448 | 2.670 | -0.0041 | No | ||
| 9 | SOD1 | 2924 | 2.241 | -0.0198 | No | ||
| 10 | CENPF | 3104 | 2.102 | -0.0201 | No | ||
| 11 | FOXL2 | 3221 | 2.015 | -0.0174 | No | ||
| 12 | GATA6 | 4291 | 1.432 | -0.0687 | No | ||
| 13 | MYF6 | 4749 | 1.257 | -0.0878 | No | ||
| 14 | TNNI3 | 5122 | 1.132 | -0.1029 | No | ||
| 15 | MYH6 | 5130 | 1.131 | -0.0982 | No | ||
| 16 | MYF5 | 6040 | 0.856 | -0.1435 | No | ||
| 17 | MYOZ1 | 6068 | 0.848 | -0.1412 | No | ||
| 18 | CHRNA1 | 6293 | 0.789 | -0.1497 | No | ||
| 19 | ITGA11 | 6514 | 0.736 | -0.1583 | No | ||
| 20 | MRAS | 6973 | 0.612 | -0.1803 | No | ||
| 21 | ITGA7 | 7388 | 0.516 | -0.2004 | No | ||
| 22 | IGF1 | 7424 | 0.506 | -0.2000 | No | ||
| 23 | ACTA1 | 7930 | 0.374 | -0.2256 | No | ||
| 24 | MYOD1 | 8054 | 0.344 | -0.2307 | No | ||
| 25 | ADAM12 | 8127 | 0.326 | -0.2332 | No | ||
| 26 | UTRN | 8171 | 0.311 | -0.2341 | No | ||
| 27 | ITGB1BP2 | 8378 | 0.261 | -0.2441 | No | ||
| 28 | UBB | 8398 | 0.256 | -0.2439 | No | ||
| 29 | TAGLN | 8434 | 0.250 | -0.2447 | No | ||
| 30 | IGFBP3 | 8459 | 0.244 | -0.2449 | No | ||
| 31 | HBEGF | 8652 | 0.195 | -0.2544 | No | ||
| 32 | COL4A4 | 8687 | 0.187 | -0.2554 | No | ||
| 33 | ACHE | 9716 | -0.075 | -0.3106 | No | ||
| 34 | CACNB2 | 9920 | -0.134 | -0.3209 | No | ||
| 35 | SGCE | 10226 | -0.205 | -0.3365 | No | ||
| 36 | BOC | 10430 | -0.245 | -0.3463 | No | ||
| 37 | GYLTL1B | 10463 | -0.252 | -0.3470 | No | ||
| 38 | MKL2 | 10575 | -0.279 | -0.3517 | No | ||
| 39 | CACNA1H | 10670 | -0.301 | -0.3554 | No | ||
| 40 | CAPN3 | 10948 | -0.373 | -0.3687 | No | ||
| 41 | FHL3 | 11018 | -0.389 | -0.3707 | No | ||
| 42 | LARGE | 11047 | -0.396 | -0.3705 | No | ||
| 43 | MUSK | 11101 | -0.408 | -0.3715 | No | ||
| 44 | LAMA2 | 11751 | -0.576 | -0.4040 | No | ||
| 45 | SPEG | 12127 | -0.683 | -0.4212 | No | ||
| 46 | MYH11 | 12173 | -0.695 | -0.4205 | No | ||
| 47 | EVC | 12564 | -0.819 | -0.4379 | No | ||
| 48 | SIX1 | 12740 | -0.871 | -0.4435 | No | ||
| 49 | CTF1 | 12757 | -0.876 | -0.4405 | No | ||
| 50 | SGCB | 12971 | -0.934 | -0.4478 | No | ||
| 51 | TEAD4 | 13456 | -1.110 | -0.4690 | No | ||
| 52 | MYBPC3 | 13878 | -1.275 | -0.4861 | No | ||
| 53 | CXCL10 | 13979 | -1.314 | -0.4856 | No | ||
| 54 | TBX3 | 13984 | -1.316 | -0.4800 | No | ||
| 55 | KCNH1 | 14558 | -1.548 | -0.5040 | No | ||
| 56 | SGCA | 14658 | -1.604 | -0.5023 | No | ||
| 57 | NF1 | 14957 | -1.771 | -0.5105 | No | ||
| 58 | MYH3 | 15194 | -1.931 | -0.5146 | No | ||
| 59 | EMD | 15521 | -2.178 | -0.5225 | Yes | ||
| 60 | MYH7 | 15533 | -2.189 | -0.5134 | Yes | ||
| 61 | TTN | 15564 | -2.218 | -0.5052 | Yes | ||
| 62 | SGCD | 15684 | -2.322 | -0.5013 | Yes | ||
| 63 | MEF2D | 15700 | -2.341 | -0.4917 | Yes | ||
| 64 | COL6A3 | 15807 | -2.440 | -0.4866 | Yes | ||
| 65 | CSRP3 | 15856 | -2.487 | -0.4781 | Yes | ||
| 66 | COL5A3 | 16099 | -2.776 | -0.4788 | Yes | ||
| 67 | SGCG | 16535 | -3.442 | -0.4870 | Yes | ||
| 68 | MTM1 | 16746 | -3.811 | -0.4814 | Yes | ||
| 69 | TAZ | 16750 | -3.820 | -0.4646 | Yes | ||
| 70 | MEF2B | 16805 | -3.929 | -0.4501 | Yes | ||
| 71 | HDAC7A | 17077 | -4.617 | -0.4442 | Yes | ||
| 72 | NOTCH1 | 17098 | -4.678 | -0.4245 | Yes | ||
| 73 | SRI | 17247 | -5.079 | -0.4099 | Yes | ||
| 74 | HDAC9 | 17284 | -5.180 | -0.3888 | Yes | ||
| 75 | MBNL1 | 17291 | -5.195 | -0.3661 | Yes | ||
| 76 | MYL6 | 17464 | -5.773 | -0.3497 | Yes | ||
| 77 | SMTN | 17471 | -5.794 | -0.3243 | Yes | ||
| 78 | TGFB1 | 17710 | -6.583 | -0.3079 | Yes | ||
| 79 | HDAC5 | 17766 | -6.776 | -0.2807 | Yes | ||
| 80 | SIRT2 | 17770 | -6.788 | -0.2508 | Yes | ||
| 81 | MEF2A | 17935 | -7.632 | -0.2257 | Yes | ||
| 82 | BMP4 | 18329 | -11.024 | -0.1979 | Yes | ||
| 83 | TCF12 | 18338 | -11.172 | -0.1487 | Yes | ||
| 84 | VAMP5 | 18340 | -11.182 | -0.0991 | Yes | ||
| 85 | MEF2C | 18390 | -12.088 | -0.0480 | Yes | ||
| 86 | POU6F1 | 18456 | -13.547 | 0.0086 | Yes |