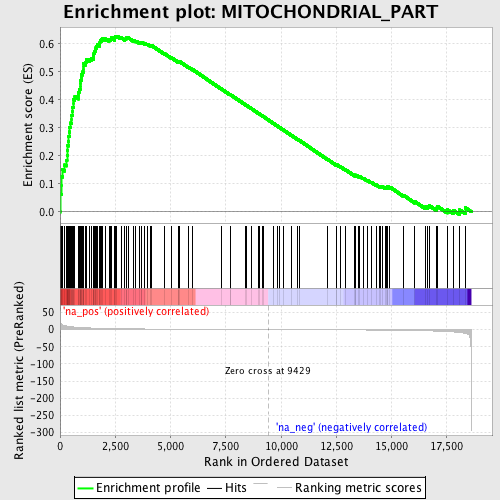
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.6277464 |
| Normalized Enrichment Score (NES) | 2.1843734 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TOMM22 | 4 | 34.032 | 0.0652 | Yes | ||
| 2 | MRPS10 | 51 | 16.878 | 0.0952 | Yes | ||
| 3 | TIMM13 | 56 | 16.536 | 0.1268 | Yes | ||
| 4 | TIMM17A | 100 | 14.118 | 0.1517 | Yes | ||
| 5 | MRPL12 | 207 | 11.350 | 0.1678 | Yes | ||
| 6 | TIMM10 | 277 | 10.400 | 0.1840 | Yes | ||
| 7 | ATP5G1 | 313 | 9.912 | 0.2012 | Yes | ||
| 8 | TOMM34 | 336 | 9.744 | 0.2188 | Yes | ||
| 9 | VDAC1 | 348 | 9.561 | 0.2366 | Yes | ||
| 10 | MRPL23 | 372 | 9.348 | 0.2533 | Yes | ||
| 11 | ABCF2 | 393 | 9.220 | 0.2699 | Yes | ||
| 12 | SUPV3L1 | 422 | 8.971 | 0.2857 | Yes | ||
| 13 | MRPS15 | 437 | 8.862 | 0.3020 | Yes | ||
| 14 | MRPS12 | 456 | 8.687 | 0.3177 | Yes | ||
| 15 | BCKDHB | 524 | 8.113 | 0.3297 | Yes | ||
| 16 | MRPS18A | 529 | 8.091 | 0.3450 | Yes | ||
| 17 | BCS1L | 551 | 7.924 | 0.3592 | Yes | ||
| 18 | IMMT | 562 | 7.849 | 0.3737 | Yes | ||
| 19 | NDUFS3 | 597 | 7.622 | 0.3865 | Yes | ||
| 20 | BAX | 601 | 7.598 | 0.4010 | Yes | ||
| 21 | PHB2 | 669 | 7.205 | 0.4112 | Yes | ||
| 22 | HTRA2 | 820 | 6.535 | 0.4157 | Yes | ||
| 23 | PMPCA | 831 | 6.494 | 0.4276 | Yes | ||
| 24 | TIMM50 | 861 | 6.352 | 0.4383 | Yes | ||
| 25 | TIMM44 | 908 | 6.136 | 0.4476 | Yes | ||
| 26 | PIN4 | 912 | 6.131 | 0.4592 | Yes | ||
| 27 | SLC25A22 | 918 | 6.111 | 0.4707 | Yes | ||
| 28 | MRPL55 | 960 | 5.951 | 0.4799 | Yes | ||
| 29 | ATP5G3 | 973 | 5.901 | 0.4906 | Yes | ||
| 30 | ABCB6 | 1010 | 5.791 | 0.4998 | Yes | ||
| 31 | ATP5J | 1045 | 5.629 | 0.5088 | Yes | ||
| 32 | ALDH4A1 | 1050 | 5.608 | 0.5194 | Yes | ||
| 33 | CYCS | 1054 | 5.594 | 0.5300 | Yes | ||
| 34 | NDUFA9 | 1161 | 5.240 | 0.5343 | Yes | ||
| 35 | MRPL40 | 1178 | 5.182 | 0.5434 | Yes | ||
| 36 | POLG2 | 1323 | 4.772 | 0.5448 | Yes | ||
| 37 | MRPS28 | 1407 | 4.500 | 0.5490 | Yes | ||
| 38 | ETFA | 1508 | 4.259 | 0.5518 | Yes | ||
| 39 | OPA1 | 1514 | 4.248 | 0.5597 | Yes | ||
| 40 | SLC25A15 | 1528 | 4.219 | 0.5671 | Yes | ||
| 41 | PPOX | 1571 | 4.125 | 0.5728 | Yes | ||
| 42 | UQCRB | 1590 | 4.081 | 0.5796 | Yes | ||
| 43 | BCL2 | 1621 | 4.010 | 0.5857 | Yes | ||
| 44 | AIFM2 | 1651 | 3.948 | 0.5918 | Yes | ||
| 45 | ATP5G2 | 1670 | 3.916 | 0.5983 | Yes | ||
| 46 | TIMM8B | 1789 | 3.694 | 0.5990 | Yes | ||
| 47 | NDUFS7 | 1799 | 3.663 | 0.6056 | Yes | ||
| 48 | ATP5O | 1809 | 3.641 | 0.6121 | Yes | ||
| 49 | ETFB | 1865 | 3.519 | 0.6159 | Yes | ||
| 50 | MRPL10 | 1929 | 3.412 | 0.6191 | Yes | ||
| 51 | NFS1 | 2066 | 3.195 | 0.6179 | Yes | ||
| 52 | MRPS24 | 2214 | 2.980 | 0.6156 | Yes | ||
| 53 | ACADM | 2287 | 2.876 | 0.6173 | Yes | ||
| 54 | DNAJA3 | 2305 | 2.857 | 0.6219 | Yes | ||
| 55 | ACN9 | 2466 | 2.656 | 0.6183 | Yes | ||
| 56 | SLC25A3 | 2473 | 2.650 | 0.6231 | Yes | ||
| 57 | ATP5A1 | 2484 | 2.635 | 0.6276 | Yes | ||
| 58 | MRPS11 | 2573 | 2.547 | 0.6277 | Yes | ||
| 59 | NDUFV1 | 2757 | 2.385 | 0.6224 | No | ||
| 60 | TFB2M | 2935 | 2.233 | 0.6172 | No | ||
| 61 | RHOT1 | 2982 | 2.194 | 0.6189 | No | ||
| 62 | BNIP3 | 3011 | 2.164 | 0.6215 | No | ||
| 63 | SDHD | 3074 | 2.123 | 0.6223 | No | ||
| 64 | NDUFAB1 | 3303 | 1.944 | 0.6137 | No | ||
| 65 | MRPS18C | 3433 | 1.855 | 0.6103 | No | ||
| 66 | MRPS35 | 3597 | 1.763 | 0.6048 | No | ||
| 67 | SURF1 | 3673 | 1.727 | 0.6041 | No | ||
| 68 | NDUFS1 | 3691 | 1.713 | 0.6065 | No | ||
| 69 | MRPL51 | 3835 | 1.637 | 0.6019 | No | ||
| 70 | TIMM9 | 3938 | 1.586 | 0.5994 | No | ||
| 71 | SLC25A1 | 4080 | 1.521 | 0.5947 | No | ||
| 72 | MFN2 | 4149 | 1.498 | 0.5939 | No | ||
| 73 | ATP5D | 4742 | 1.261 | 0.5643 | No | ||
| 74 | MPV17 | 5042 | 1.158 | 0.5504 | No | ||
| 75 | TIMM23 | 5360 | 1.063 | 0.5353 | No | ||
| 76 | HCCS | 5364 | 1.059 | 0.5372 | No | ||
| 77 | CENTA2 | 5397 | 1.043 | 0.5374 | No | ||
| 78 | MTX2 | 5812 | 0.922 | 0.5168 | No | ||
| 79 | DBT | 6002 | 0.867 | 0.5083 | No | ||
| 80 | VDAC3 | 7287 | 0.539 | 0.4399 | No | ||
| 81 | MRPS21 | 7690 | 0.433 | 0.4189 | No | ||
| 82 | MRPS16 | 7717 | 0.427 | 0.4184 | No | ||
| 83 | VDAC2 | 8383 | 0.260 | 0.3829 | No | ||
| 84 | MRPS22 | 8435 | 0.250 | 0.3806 | No | ||
| 85 | PITRM1 | 8659 | 0.193 | 0.3689 | No | ||
| 86 | SDHA | 8964 | 0.121 | 0.3527 | No | ||
| 87 | COX15 | 9007 | 0.110 | 0.3506 | No | ||
| 88 | NDUFA2 | 9163 | 0.065 | 0.3424 | No | ||
| 89 | ATP5F1 | 9182 | 0.061 | 0.3415 | No | ||
| 90 | ATP5E | 9671 | -0.065 | 0.3153 | No | ||
| 91 | UQCRC1 | 9821 | -0.107 | 0.3074 | No | ||
| 92 | GRPEL1 | 9916 | -0.132 | 0.3026 | No | ||
| 93 | CS | 10116 | -0.181 | 0.2922 | No | ||
| 94 | NDUFA1 | 10478 | -0.256 | 0.2731 | No | ||
| 95 | HSD3B1 | 10731 | -0.316 | 0.2601 | No | ||
| 96 | ATP5B | 10820 | -0.337 | 0.2560 | No | ||
| 97 | ALAS2 | 12107 | -0.678 | 0.1877 | No | ||
| 98 | MRPL52 | 12507 | -0.802 | 0.1677 | No | ||
| 99 | MAOB | 12519 | -0.806 | 0.1687 | No | ||
| 100 | RAB11FIP5 | 12680 | -0.852 | 0.1616 | No | ||
| 101 | BCKDK | 12908 | -0.918 | 0.1511 | No | ||
| 102 | NDUFA13 | 13334 | -1.064 | 0.1302 | No | ||
| 103 | CASP7 | 13374 | -1.077 | 0.1302 | No | ||
| 104 | PHB | 13379 | -1.079 | 0.1320 | No | ||
| 105 | ATP5C1 | 13518 | -1.135 | 0.1267 | No | ||
| 106 | NDUFS2 | 13555 | -1.152 | 0.1270 | No | ||
| 107 | HSD3B2 | 13713 | -1.218 | 0.1208 | No | ||
| 108 | MRPS36 | 13908 | -1.288 | 0.1128 | No | ||
| 109 | OGDH | 14082 | -1.354 | 0.1061 | No | ||
| 110 | CASQ1 | 14302 | -1.438 | 0.0970 | No | ||
| 111 | NDUFA6 | 14477 | -1.512 | 0.0905 | No | ||
| 112 | NDUFS4 | 14496 | -1.518 | 0.0924 | No | ||
| 113 | COX6B2 | 14577 | -1.555 | 0.0911 | No | ||
| 114 | GATM | 14715 | -1.633 | 0.0868 | No | ||
| 115 | UCP3 | 14755 | -1.656 | 0.0879 | No | ||
| 116 | RAF1 | 14806 | -1.685 | 0.0885 | No | ||
| 117 | HADHB | 14823 | -1.696 | 0.0909 | No | ||
| 118 | UQCRH | 14930 | -1.757 | 0.0885 | No | ||
| 119 | NDUFS8 | 15528 | -2.187 | 0.0604 | No | ||
| 120 | BCKDHA | 16060 | -2.729 | 0.0369 | No | ||
| 121 | NNT | 16532 | -3.439 | 0.0181 | No | ||
| 122 | RHOT2 | 16625 | -3.606 | 0.0200 | No | ||
| 123 | MRPL32 | 16715 | -3.758 | 0.0225 | No | ||
| 124 | ABCB7 | 17029 | -4.480 | 0.0141 | No | ||
| 125 | NR3C1 | 17064 | -4.576 | 0.0211 | No | ||
| 126 | FIS1 | 17525 | -6.012 | 0.0078 | No | ||
| 127 | SLC25A11 | 17790 | -6.885 | 0.0068 | No | ||
| 128 | MCL1 | 18083 | -8.510 | 0.0073 | No | ||
| 129 | TIMM17B | 18337 | -11.148 | 0.0151 | No |
