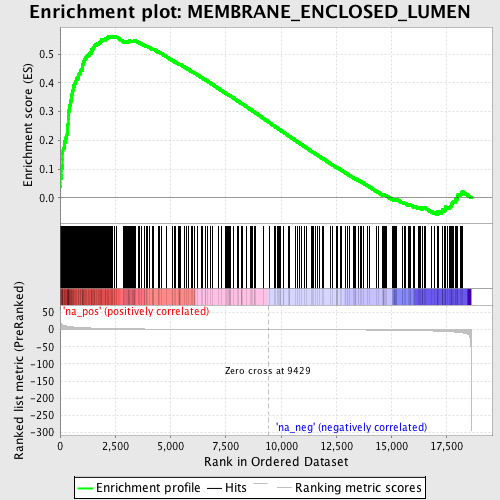
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MEMBRANE_ENCLOSED_LUMEN |
| Enrichment Score (ES) | 0.5629499 |
| Normalized Enrichment Score (NES) | 2.1699412 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDKN2A | 0 | 53.800 | 0.0404 | Yes | ||
| 2 | IKBKAP | 30 | 18.923 | 0.0530 | Yes | ||
| 3 | NOL1 | 31 | 18.917 | 0.0672 | Yes | ||
| 4 | CIRH1A | 45 | 17.130 | 0.0793 | Yes | ||
| 5 | MRPS10 | 51 | 16.878 | 0.0917 | Yes | ||
| 6 | GNL3 | 53 | 16.799 | 0.1043 | Yes | ||
| 7 | EXOSC1 | 90 | 14.634 | 0.1133 | Yes | ||
| 8 | NOL6 | 97 | 14.194 | 0.1236 | Yes | ||
| 9 | MYBBP1A | 98 | 14.156 | 0.1342 | Yes | ||
| 10 | NOLA2 | 103 | 13.964 | 0.1445 | Yes | ||
| 11 | RPP38 | 105 | 13.842 | 0.1548 | Yes | ||
| 12 | RRP9 | 128 | 13.247 | 0.1636 | Yes | ||
| 13 | SDAD1 | 131 | 13.197 | 0.1734 | Yes | ||
| 14 | U2AF1 | 184 | 11.817 | 0.1794 | Yes | ||
| 15 | LYAR | 201 | 11.440 | 0.1871 | Yes | ||
| 16 | MRPL12 | 207 | 11.350 | 0.1954 | Yes | ||
| 17 | DDX21 | 240 | 10.796 | 0.2017 | Yes | ||
| 18 | CPSF1 | 260 | 10.576 | 0.2086 | Yes | ||
| 19 | MKI67IP | 293 | 10.191 | 0.2145 | Yes | ||
| 20 | NOLA3 | 310 | 9.971 | 0.2211 | Yes | ||
| 21 | GTF2H4 | 314 | 9.906 | 0.2284 | Yes | ||
| 22 | POLR3H | 329 | 9.824 | 0.2350 | Yes | ||
| 23 | WDHD1 | 332 | 9.784 | 0.2422 | Yes | ||
| 24 | CHAF1B | 339 | 9.719 | 0.2492 | Yes | ||
| 25 | GEMIN5 | 352 | 9.516 | 0.2557 | Yes | ||
| 26 | ELP3 | 360 | 9.459 | 0.2624 | Yes | ||
| 27 | PPARGC1B | 362 | 9.444 | 0.2694 | Yes | ||
| 28 | MPG | 363 | 9.439 | 0.2765 | Yes | ||
| 29 | MRPL23 | 372 | 9.348 | 0.2831 | Yes | ||
| 30 | NOLC1 | 378 | 9.329 | 0.2898 | Yes | ||
| 31 | NCL | 390 | 9.246 | 0.2961 | Yes | ||
| 32 | XRN2 | 395 | 9.212 | 0.3028 | Yes | ||
| 33 | RUVBL1 | 414 | 9.053 | 0.3087 | Yes | ||
| 34 | SUPV3L1 | 422 | 8.971 | 0.3150 | Yes | ||
| 35 | MRPS15 | 437 | 8.862 | 0.3209 | Yes | ||
| 36 | MRPS12 | 456 | 8.687 | 0.3264 | Yes | ||
| 37 | RUVBL2 | 469 | 8.535 | 0.3322 | Yes | ||
| 38 | CDK8 | 483 | 8.450 | 0.3378 | Yes | ||
| 39 | ERCC8 | 506 | 8.311 | 0.3428 | Yes | ||
| 40 | POLR1E | 522 | 8.137 | 0.3481 | Yes | ||
| 41 | BCKDHB | 524 | 8.113 | 0.3542 | Yes | ||
| 42 | MRPS18A | 529 | 8.091 | 0.3600 | Yes | ||
| 43 | MORF4L2 | 564 | 7.838 | 0.3640 | Yes | ||
| 44 | POLR2J | 575 | 7.790 | 0.3693 | Yes | ||
| 45 | CHAF1A | 578 | 7.784 | 0.3751 | Yes | ||
| 46 | GEMIN6 | 598 | 7.605 | 0.3797 | Yes | ||
| 47 | RPA1 | 599 | 7.604 | 0.3854 | Yes | ||
| 48 | NSUN2 | 603 | 7.571 | 0.3910 | Yes | ||
| 49 | ASH2L | 651 | 7.277 | 0.3939 | Yes | ||
| 50 | GTF2H1 | 671 | 7.198 | 0.3982 | Yes | ||
| 51 | MCM7 | 698 | 7.027 | 0.4021 | Yes | ||
| 52 | GEMIN4 | 708 | 6.976 | 0.4068 | Yes | ||
| 53 | HDAC6 | 720 | 6.919 | 0.4114 | Yes | ||
| 54 | PARP1 | 742 | 6.797 | 0.4154 | Yes | ||
| 55 | COIL | 780 | 6.678 | 0.4183 | Yes | ||
| 56 | NOL5A | 817 | 6.547 | 0.4213 | Yes | ||
| 57 | HTRA2 | 820 | 6.535 | 0.4261 | Yes | ||
| 58 | NARG1 | 821 | 6.535 | 0.4310 | Yes | ||
| 59 | TIMM50 | 861 | 6.352 | 0.4336 | Yes | ||
| 60 | TIMM44 | 908 | 6.136 | 0.4357 | Yes | ||
| 61 | PIN4 | 912 | 6.131 | 0.4401 | Yes | ||
| 62 | POLI | 937 | 6.030 | 0.4434 | Yes | ||
| 63 | MRPL55 | 960 | 5.951 | 0.4466 | Yes | ||
| 64 | HDAC10 | 998 | 5.821 | 0.4490 | Yes | ||
| 65 | GTF3C3 | 1005 | 5.800 | 0.4530 | Yes | ||
| 66 | MRE11A | 1007 | 5.796 | 0.4573 | Yes | ||
| 67 | SMC3 | 1015 | 5.758 | 0.4612 | Yes | ||
| 68 | TRIM27 | 1016 | 5.745 | 0.4655 | Yes | ||
| 69 | ALDH4A1 | 1050 | 5.608 | 0.4679 | Yes | ||
| 70 | CYCS | 1054 | 5.594 | 0.4720 | Yes | ||
| 71 | GTF2F1 | 1058 | 5.591 | 0.4760 | Yes | ||
| 72 | NUP54 | 1098 | 5.452 | 0.4780 | Yes | ||
| 73 | NAP1L1 | 1102 | 5.432 | 0.4819 | Yes | ||
| 74 | PTBP1 | 1135 | 5.338 | 0.4841 | Yes | ||
| 75 | POLR2E | 1154 | 5.265 | 0.4871 | Yes | ||
| 76 | MRPL40 | 1178 | 5.182 | 0.4897 | Yes | ||
| 77 | WDR4 | 1182 | 5.172 | 0.4934 | Yes | ||
| 78 | IVNS1ABP | 1236 | 5.009 | 0.4943 | Yes | ||
| 79 | CLOCK | 1275 | 4.902 | 0.4959 | Yes | ||
| 80 | SURF6 | 1277 | 4.897 | 0.4995 | Yes | ||
| 81 | POLG2 | 1323 | 4.772 | 0.5007 | Yes | ||
| 82 | KRR1 | 1328 | 4.756 | 0.5040 | Yes | ||
| 83 | RPP30 | 1367 | 4.626 | 0.5054 | Yes | ||
| 84 | SMARCC1 | 1403 | 4.506 | 0.5069 | Yes | ||
| 85 | MRPS28 | 1407 | 4.500 | 0.5101 | Yes | ||
| 86 | SNAPC4 | 1408 | 4.499 | 0.5135 | Yes | ||
| 87 | SRP68 | 1430 | 4.450 | 0.5156 | Yes | ||
| 88 | POP1 | 1474 | 4.345 | 0.5166 | Yes | ||
| 89 | ILF2 | 1483 | 4.306 | 0.5193 | Yes | ||
| 90 | ETFA | 1508 | 4.259 | 0.5212 | Yes | ||
| 91 | OPA1 | 1514 | 4.248 | 0.5241 | Yes | ||
| 92 | HDAC1 | 1535 | 4.202 | 0.5262 | Yes | ||
| 93 | MPHOSPH1 | 1536 | 4.202 | 0.5294 | Yes | ||
| 94 | NAP1L4 | 1557 | 4.142 | 0.5314 | Yes | ||
| 95 | TCOF1 | 1588 | 4.083 | 0.5328 | Yes | ||
| 96 | CHEK2 | 1647 | 3.955 | 0.5326 | Yes | ||
| 97 | SLC29A2 | 1656 | 3.941 | 0.5351 | Yes | ||
| 98 | HDAC2 | 1672 | 3.914 | 0.5372 | Yes | ||
| 99 | SMARCE1 | 1728 | 3.808 | 0.5371 | Yes | ||
| 100 | GTF3C1 | 1751 | 3.763 | 0.5387 | Yes | ||
| 101 | PRPF31 | 1783 | 3.699 | 0.5398 | Yes | ||
| 102 | POLQ | 1796 | 3.669 | 0.5419 | Yes | ||
| 103 | POLR2A | 1817 | 3.619 | 0.5435 | Yes | ||
| 104 | HMGA1 | 1826 | 3.607 | 0.5458 | Yes | ||
| 105 | ETFB | 1865 | 3.519 | 0.5463 | Yes | ||
| 106 | SFRS2 | 1866 | 3.517 | 0.5490 | Yes | ||
| 107 | FOXF2 | 1867 | 3.516 | 0.5516 | Yes | ||
| 108 | MRPL10 | 1929 | 3.412 | 0.5508 | Yes | ||
| 109 | RPS19 | 1945 | 3.390 | 0.5526 | Yes | ||
| 110 | UBTF | 1999 | 3.301 | 0.5521 | Yes | ||
| 111 | DDX11 | 2055 | 3.212 | 0.5515 | Yes | ||
| 112 | EED | 2064 | 3.200 | 0.5535 | Yes | ||
| 113 | NFS1 | 2066 | 3.195 | 0.5559 | Yes | ||
| 114 | ZNF638 | 2111 | 3.127 | 0.5558 | Yes | ||
| 115 | SMARCD3 | 2149 | 3.075 | 0.5561 | Yes | ||
| 116 | TAF6 | 2153 | 3.072 | 0.5582 | Yes | ||
| 117 | FUSIP1 | 2172 | 3.035 | 0.5595 | Yes | ||
| 118 | TBP | 2197 | 3.003 | 0.5605 | Yes | ||
| 119 | MRPS24 | 2214 | 2.980 | 0.5618 | Yes | ||
| 120 | ACADM | 2287 | 2.876 | 0.5600 | Yes | ||
| 121 | DNAJA3 | 2305 | 2.857 | 0.5612 | Yes | ||
| 122 | TOP2A | 2322 | 2.831 | 0.5625 | Yes | ||
| 123 | EXOSC10 | 2383 | 2.742 | 0.5613 | Yes | ||
| 124 | POLR3C | 2391 | 2.732 | 0.5629 | Yes | ||
| 125 | KPNA2 | 2441 | 2.674 | 0.5623 | Yes | ||
| 126 | ACN9 | 2466 | 2.656 | 0.5629 | Yes | ||
| 127 | MRPS11 | 2573 | 2.547 | 0.5591 | No | ||
| 128 | NR6A1 | 2881 | 2.279 | 0.5440 | No | ||
| 129 | MED4 | 2885 | 2.275 | 0.5455 | No | ||
| 130 | TFB2M | 2935 | 2.233 | 0.5445 | No | ||
| 131 | GTF3C4 | 2979 | 2.196 | 0.5438 | No | ||
| 132 | BNIP3 | 3011 | 2.164 | 0.5438 | No | ||
| 133 | ARID1A | 3070 | 2.125 | 0.5422 | No | ||
| 134 | FMR1 | 3089 | 2.112 | 0.5428 | No | ||
| 135 | CENPF | 3104 | 2.102 | 0.5436 | No | ||
| 136 | CPSF3 | 3114 | 2.093 | 0.5447 | No | ||
| 137 | ACTL6A | 3124 | 2.086 | 0.5457 | No | ||
| 138 | GTF3C5 | 3139 | 2.074 | 0.5465 | No | ||
| 139 | TAF5 | 3186 | 2.040 | 0.5455 | No | ||
| 140 | HDAC3 | 3242 | 1.996 | 0.5440 | No | ||
| 141 | EMG1 | 3269 | 1.974 | 0.5441 | No | ||
| 142 | GLIS2 | 3282 | 1.966 | 0.5449 | No | ||
| 143 | NDUFAB1 | 3303 | 1.944 | 0.5453 | No | ||
| 144 | PHF12 | 3318 | 1.933 | 0.5460 | No | ||
| 145 | NPM1 | 3360 | 1.903 | 0.5452 | No | ||
| 146 | MED6 | 3366 | 1.899 | 0.5463 | No | ||
| 147 | TAF4 | 3376 | 1.891 | 0.5472 | No | ||
| 148 | MRPS18C | 3433 | 1.855 | 0.5456 | No | ||
| 149 | LRCH4 | 3549 | 1.788 | 0.5406 | No | ||
| 150 | MRPS35 | 3597 | 1.763 | 0.5394 | No | ||
| 151 | NDUFS1 | 3691 | 1.713 | 0.5356 | No | ||
| 152 | NF2 | 3819 | 1.643 | 0.5299 | No | ||
| 153 | MRPL51 | 3835 | 1.637 | 0.5303 | No | ||
| 154 | IFI16 | 3912 | 1.596 | 0.5273 | No | ||
| 155 | EDF1 | 3913 | 1.596 | 0.5285 | No | ||
| 156 | RPL3 | 3963 | 1.574 | 0.5270 | No | ||
| 157 | RSF1 | 4043 | 1.537 | 0.5238 | No | ||
| 158 | ARID4A | 4176 | 1.483 | 0.5177 | No | ||
| 159 | HNRPK | 4194 | 1.474 | 0.5179 | No | ||
| 160 | DDX47 | 4212 | 1.467 | 0.5181 | No | ||
| 161 | NUDT21 | 4245 | 1.454 | 0.5174 | No | ||
| 162 | HIF1A | 4444 | 1.376 | 0.5076 | No | ||
| 163 | TDG | 4455 | 1.373 | 0.5081 | No | ||
| 164 | ASNA1 | 4514 | 1.346 | 0.5059 | No | ||
| 165 | PRM2 | 4581 | 1.322 | 0.5033 | No | ||
| 166 | TAF9 | 4814 | 1.226 | 0.4916 | No | ||
| 167 | DNMT3A | 5090 | 1.145 | 0.4774 | No | ||
| 168 | RPP40 | 5105 | 1.139 | 0.4775 | No | ||
| 169 | GCM1 | 5197 | 1.108 | 0.4733 | No | ||
| 170 | SAP18 | 5230 | 1.100 | 0.4724 | No | ||
| 171 | TIMM23 | 5360 | 1.063 | 0.4661 | No | ||
| 172 | PAX8 | 5386 | 1.051 | 0.4656 | No | ||
| 173 | NCR1 | 5395 | 1.044 | 0.4659 | No | ||
| 174 | OIP5 | 5415 | 1.038 | 0.4657 | No | ||
| 175 | DDX56 | 5449 | 1.028 | 0.4646 | No | ||
| 176 | FIGLA | 5632 | 0.976 | 0.4554 | No | ||
| 177 | SMARCA5 | 5702 | 0.953 | 0.4523 | No | ||
| 178 | NKX2-5 | 5827 | 0.918 | 0.4462 | No | ||
| 179 | NPAS2 | 5966 | 0.878 | 0.4394 | No | ||
| 180 | DBT | 6002 | 0.867 | 0.4381 | No | ||
| 181 | PITX2 | 6070 | 0.848 | 0.4351 | No | ||
| 182 | MBTPS1 | 6090 | 0.841 | 0.4347 | No | ||
| 183 | INTS6 | 6219 | 0.809 | 0.4283 | No | ||
| 184 | ACTB | 6384 | 0.769 | 0.4199 | No | ||
| 185 | RBBP5 | 6454 | 0.749 | 0.4167 | No | ||
| 186 | TAF12 | 6565 | 0.719 | 0.4112 | No | ||
| 187 | POP7 | 6593 | 0.711 | 0.4102 | No | ||
| 188 | ERCC3 | 6652 | 0.699 | 0.4076 | No | ||
| 189 | OGG1 | 6817 | 0.655 | 0.3991 | No | ||
| 190 | TOP1 | 6905 | 0.631 | 0.3948 | No | ||
| 191 | SHFM1 | 7190 | 0.562 | 0.3797 | No | ||
| 192 | TAF11 | 7313 | 0.533 | 0.3735 | No | ||
| 193 | GTF3C2 | 7483 | 0.488 | 0.3646 | No | ||
| 194 | RCN1 | 7520 | 0.478 | 0.3630 | No | ||
| 195 | TOP2B | 7594 | 0.461 | 0.3593 | No | ||
| 196 | HSP90B1 | 7604 | 0.458 | 0.3592 | No | ||
| 197 | RTCD1 | 7672 | 0.439 | 0.3558 | No | ||
| 198 | UBE2I | 7676 | 0.438 | 0.3560 | No | ||
| 199 | MRPS21 | 7690 | 0.433 | 0.3556 | No | ||
| 200 | MRPS16 | 7717 | 0.427 | 0.3545 | No | ||
| 201 | NCOA6 | 7835 | 0.394 | 0.3484 | No | ||
| 202 | HIPK2 | 8023 | 0.353 | 0.3385 | No | ||
| 203 | RPL36 | 8077 | 0.339 | 0.3358 | No | ||
| 204 | MED12 | 8198 | 0.304 | 0.3295 | No | ||
| 205 | NFYC | 8214 | 0.299 | 0.3289 | No | ||
| 206 | DDX24 | 8224 | 0.296 | 0.3286 | No | ||
| 207 | SAP30 | 8258 | 0.288 | 0.3270 | No | ||
| 208 | MRPS22 | 8435 | 0.250 | 0.3176 | No | ||
| 209 | SMARCB1 | 8603 | 0.208 | 0.3086 | No | ||
| 210 | PITRM1 | 8659 | 0.193 | 0.3057 | No | ||
| 211 | XPO1 | 8722 | 0.178 | 0.3025 | No | ||
| 212 | KIN | 8791 | 0.163 | 0.2989 | No | ||
| 213 | NUP98 | 8813 | 0.159 | 0.2979 | No | ||
| 214 | SP100 | 8820 | 0.157 | 0.2977 | No | ||
| 215 | ATP5F1 | 9182 | 0.061 | 0.2780 | No | ||
| 216 | EPAS1 | 9487 | -0.019 | 0.2613 | No | ||
| 217 | POLA1 | 9723 | -0.077 | 0.2486 | No | ||
| 218 | TEP1 | 9743 | -0.082 | 0.2476 | No | ||
| 219 | PMF1 | 9819 | -0.107 | 0.2436 | No | ||
| 220 | POLH | 9902 | -0.129 | 0.2392 | No | ||
| 221 | GRPEL1 | 9916 | -0.132 | 0.2386 | No | ||
| 222 | FOXE3 | 9958 | -0.143 | 0.2364 | No | ||
| 223 | TOPBP1 | 9989 | -0.149 | 0.2349 | No | ||
| 224 | TAP2 | 10114 | -0.181 | 0.2282 | No | ||
| 225 | CS | 10116 | -0.181 | 0.2283 | No | ||
| 226 | NAP1L3 | 10329 | -0.226 | 0.2169 | No | ||
| 227 | ATXN1 | 10372 | -0.234 | 0.2148 | No | ||
| 228 | TFIP11 | 10382 | -0.236 | 0.2145 | No | ||
| 229 | HS3ST1 | 10631 | -0.291 | 0.2011 | No | ||
| 230 | HSD3B1 | 10731 | -0.316 | 0.1959 | No | ||
| 231 | THRAP3 | 10854 | -0.346 | 0.1895 | No | ||
| 232 | HTATIP | 10913 | -0.364 | 0.1866 | No | ||
| 233 | ATXN3 | 11069 | -0.401 | 0.1785 | No | ||
| 234 | PTF1A | 11131 | -0.416 | 0.1754 | No | ||
| 235 | NEK11 | 11382 | -0.481 | 0.1621 | No | ||
| 236 | SMAD2 | 11415 | -0.487 | 0.1607 | No | ||
| 237 | RPS7 | 11469 | -0.500 | 0.1582 | No | ||
| 238 | DNAJB9 | 11556 | -0.522 | 0.1539 | No | ||
| 239 | CALR | 11661 | -0.551 | 0.1486 | No | ||
| 240 | WRN | 11741 | -0.574 | 0.1447 | No | ||
| 241 | PHF21A | 11886 | -0.615 | 0.1373 | No | ||
| 242 | CHMP1A | 11932 | -0.628 | 0.1353 | No | ||
| 243 | EXOSC9 | 12244 | -0.714 | 0.1189 | No | ||
| 244 | DYRK1A | 12335 | -0.746 | 0.1145 | No | ||
| 245 | MRPL52 | 12507 | -0.802 | 0.1058 | No | ||
| 246 | SFRS2IP | 12521 | -0.806 | 0.1057 | No | ||
| 247 | XRCC6 | 12541 | -0.812 | 0.1052 | No | ||
| 248 | SMARCA4 | 12552 | -0.815 | 0.1053 | No | ||
| 249 | FGF18 | 12676 | -0.851 | 0.0992 | No | ||
| 250 | SMARCD1 | 12724 | -0.866 | 0.0973 | No | ||
| 251 | BCKDK | 12908 | -0.918 | 0.0880 | No | ||
| 252 | TAF6L | 13014 | -0.948 | 0.0829 | No | ||
| 253 | TXNDC4 | 13111 | -0.982 | 0.0784 | No | ||
| 254 | NAP1L2 | 13294 | -1.050 | 0.0693 | No | ||
| 255 | NDUFA13 | 13334 | -1.064 | 0.0679 | No | ||
| 256 | PHB | 13379 | -1.079 | 0.0663 | No | ||
| 257 | NOL4 | 13384 | -1.081 | 0.0669 | No | ||
| 258 | NLRP5 | 13497 | -1.126 | 0.0616 | No | ||
| 259 | ING2 | 13597 | -1.170 | 0.0571 | No | ||
| 260 | ELP4 | 13642 | -1.188 | 0.0556 | No | ||
| 261 | HSD3B2 | 13713 | -1.218 | 0.0527 | No | ||
| 262 | MRPS36 | 13908 | -1.288 | 0.0430 | No | ||
| 263 | RPL11 | 14015 | -1.328 | 0.0382 | No | ||
| 264 | CASQ1 | 14302 | -1.438 | 0.0237 | No | ||
| 265 | NFYA | 14396 | -1.477 | 0.0197 | No | ||
| 266 | ISG20 | 14584 | -1.557 | 0.0106 | No | ||
| 267 | CBX1 | 14637 | -1.593 | 0.0090 | No | ||
| 268 | NUFIP1 | 14648 | -1.599 | 0.0096 | No | ||
| 269 | TOPORS | 14649 | -1.599 | 0.0108 | No | ||
| 270 | POLR2B | 14675 | -1.616 | 0.0107 | No | ||
| 271 | GATM | 14715 | -1.633 | 0.0098 | No | ||
| 272 | NOL3 | 14789 | -1.674 | 0.0070 | No | ||
| 273 | RBM14 | 15023 | -1.811 | -0.0043 | No | ||
| 274 | ABL1 | 15100 | -1.861 | -0.0071 | No | ||
| 275 | ZNF148 | 15121 | -1.881 | -0.0068 | No | ||
| 276 | CIB1 | 15143 | -1.893 | -0.0065 | No | ||
| 277 | ELF4 | 15152 | -1.898 | -0.0055 | No | ||
| 278 | RPL35 | 15198 | -1.936 | -0.0065 | No | ||
| 279 | MYO6 | 15199 | -1.937 | -0.0051 | No | ||
| 280 | PPARGC1A | 15223 | -1.951 | -0.0049 | No | ||
| 281 | SPTBN4 | 15238 | -1.959 | -0.0042 | No | ||
| 282 | TAF7 | 15476 | -2.150 | -0.0155 | No | ||
| 283 | ZBTB16 | 15483 | -2.155 | -0.0142 | No | ||
| 284 | KLHL7 | 15588 | -2.238 | -0.0182 | No | ||
| 285 | POLR3K | 15639 | -2.284 | -0.0193 | No | ||
| 286 | ZNF346 | 15754 | -2.386 | -0.0237 | No | ||
| 287 | RBPJ | 15794 | -2.430 | -0.0240 | No | ||
| 288 | MPHOSPH10 | 15861 | -2.494 | -0.0257 | No | ||
| 289 | CCNT1 | 15870 | -2.503 | -0.0243 | No | ||
| 290 | ELL2 | 16008 | -2.674 | -0.0298 | No | ||
| 291 | MCRS1 | 16051 | -2.723 | -0.0300 | No | ||
| 292 | BCKDHA | 16060 | -2.729 | -0.0284 | No | ||
| 293 | HDAC11 | 16233 | -2.946 | -0.0356 | No | ||
| 294 | SEP15 | 16281 | -3.000 | -0.0359 | No | ||
| 295 | DEDD | 16293 | -3.024 | -0.0343 | No | ||
| 296 | HDAC8 | 16401 | -3.219 | -0.0377 | No | ||
| 297 | TAF10 | 16402 | -3.220 | -0.0353 | No | ||
| 298 | MEN1 | 16423 | -3.244 | -0.0340 | No | ||
| 299 | APTX | 16505 | -3.388 | -0.0358 | No | ||
| 300 | RB1 | 16529 | -3.437 | -0.0345 | No | ||
| 301 | SMARCD2 | 16797 | -3.912 | -0.0462 | No | ||
| 302 | TAF13 | 16957 | -4.266 | -0.0517 | No | ||
| 303 | NR3C1 | 17064 | -4.576 | -0.0540 | No | ||
| 304 | RELA | 17065 | -4.586 | -0.0506 | No | ||
| 305 | HDAC7A | 17077 | -4.617 | -0.0477 | No | ||
| 306 | ERCC5 | 17123 | -4.746 | -0.0466 | No | ||
| 307 | HDAC9 | 17284 | -5.180 | -0.0515 | No | ||
| 308 | TRPC4AP | 17286 | -5.185 | -0.0477 | No | ||
| 309 | EZH2 | 17294 | -5.203 | -0.0441 | No | ||
| 310 | DDX54 | 17300 | -5.230 | -0.0405 | No | ||
| 311 | HES6 | 17412 | -5.550 | -0.0424 | No | ||
| 312 | RBM39 | 17426 | -5.574 | -0.0389 | No | ||
| 313 | CDK9 | 17439 | -5.654 | -0.0353 | No | ||
| 314 | SMUG1 | 17441 | -5.666 | -0.0311 | No | ||
| 315 | MDM2 | 17547 | -6.062 | -0.0323 | No | ||
| 316 | NFYB | 17634 | -6.332 | -0.0323 | No | ||
| 317 | PPP1R9B | 17682 | -6.443 | -0.0300 | No | ||
| 318 | PML | 17701 | -6.528 | -0.0261 | No | ||
| 319 | YEATS4 | 17724 | -6.644 | -0.0223 | No | ||
| 320 | GEMIN7 | 17734 | -6.686 | -0.0178 | No | ||
| 321 | HDAC5 | 17766 | -6.776 | -0.0144 | No | ||
| 322 | ASF1A | 17797 | -6.919 | -0.0109 | No | ||
| 323 | GIT2 | 17890 | -7.395 | -0.0103 | No | ||
| 324 | HCLS1 | 17891 | -7.404 | -0.0048 | No | ||
| 325 | EPC1 | 17947 | -7.671 | -0.0020 | No | ||
| 326 | DEDD2 | 17983 | -7.810 | 0.0019 | No | ||
| 327 | ERCC2 | 17992 | -7.856 | 0.0074 | No | ||
| 328 | ARNTL | 18007 | -7.954 | 0.0126 | No | ||
| 329 | SMAD4 | 18108 | -8.699 | 0.0136 | No | ||
| 330 | LMO4 | 18154 | -9.070 | 0.0180 | No | ||
| 331 | TP53 | 18216 | -9.624 | 0.0219 | No |
