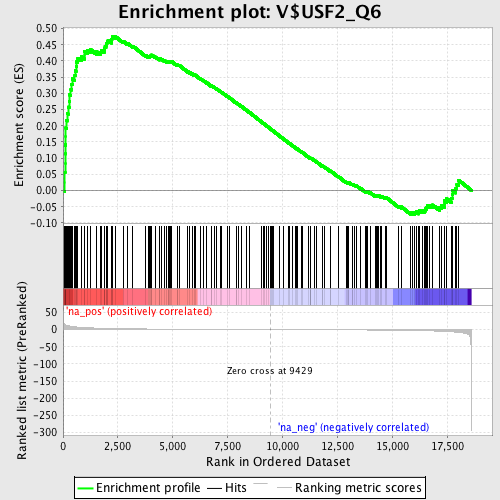
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$USF2_Q6 |
| Enrichment Score (ES) | 0.4767346 |
| Normalized Enrichment Score (NES) | 1.738325 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009097667 |
| FWER p-Value | 0.242 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FKBP11 | 82 | 14.885 | 0.0254 | Yes | ||
| 2 | B3GALT6 | 83 | 14.867 | 0.0553 | Yes | ||
| 3 | ATAD3A | 91 | 14.629 | 0.0843 | Yes | ||
| 4 | ADK | 94 | 14.400 | 0.1131 | Yes | ||
| 5 | EBNA1BP2 | 101 | 14.096 | 0.1411 | Yes | ||
| 6 | LDHA | 123 | 13.322 | 0.1667 | Yes | ||
| 7 | NUDC | 126 | 13.274 | 0.1932 | Yes | ||
| 8 | AK2 | 163 | 12.258 | 0.2159 | Yes | ||
| 9 | PPRC1 | 187 | 11.752 | 0.2382 | Yes | ||
| 10 | DPAGT1 | 230 | 10.951 | 0.2579 | Yes | ||
| 11 | TIMM10 | 277 | 10.400 | 0.2763 | Yes | ||
| 12 | CSDA | 298 | 10.136 | 0.2956 | Yes | ||
| 13 | PABPC4 | 354 | 9.488 | 0.3117 | Yes | ||
| 14 | AMPD2 | 382 | 9.297 | 0.3289 | Yes | ||
| 15 | PA2G4 | 417 | 9.021 | 0.3451 | Yes | ||
| 16 | COPS7A | 534 | 8.039 | 0.3550 | Yes | ||
| 17 | EIF3S10 | 555 | 7.895 | 0.3698 | Yes | ||
| 18 | PICALM | 593 | 7.642 | 0.3831 | Yes | ||
| 19 | ZZZ3 | 613 | 7.527 | 0.3972 | Yes | ||
| 20 | GTF2H1 | 671 | 7.198 | 0.4086 | Yes | ||
| 21 | SFXN2 | 828 | 6.502 | 0.4132 | Yes | ||
| 22 | ARL2 | 967 | 5.919 | 0.4176 | Yes | ||
| 23 | CUGBP1 | 969 | 5.911 | 0.4294 | Yes | ||
| 24 | FEN1 | 1108 | 5.410 | 0.4328 | Yes | ||
| 25 | IVNS1ABP | 1236 | 5.009 | 0.4359 | Yes | ||
| 26 | MTCH2 | 1525 | 4.226 | 0.4288 | Yes | ||
| 27 | IL15RA | 1693 | 3.875 | 0.4276 | Yes | ||
| 28 | HNRPA1 | 1726 | 3.812 | 0.4335 | Yes | ||
| 29 | CENTB5 | 1869 | 3.511 | 0.4328 | Yes | ||
| 30 | CHD4 | 1881 | 3.494 | 0.4392 | Yes | ||
| 31 | BHLHB3 | 1908 | 3.447 | 0.4448 | Yes | ||
| 32 | RHEBL1 | 1956 | 3.372 | 0.4490 | Yes | ||
| 33 | EIF4B | 1981 | 3.329 | 0.4544 | Yes | ||
| 34 | ALDH3B1 | 2003 | 3.292 | 0.4598 | Yes | ||
| 35 | PFDN2 | 2044 | 3.232 | 0.4642 | Yes | ||
| 36 | RGL1 | 2210 | 2.986 | 0.4612 | Yes | ||
| 37 | ARL3 | 2212 | 2.981 | 0.4671 | Yes | ||
| 38 | TAGLN2 | 2230 | 2.945 | 0.4721 | Yes | ||
| 39 | CCAR1 | 2254 | 2.910 | 0.4767 | Yes | ||
| 40 | POLR3C | 2391 | 2.732 | 0.4749 | No | ||
| 41 | FABP3 | 2729 | 2.411 | 0.4614 | No | ||
| 42 | TFB2M | 2935 | 2.233 | 0.4548 | No | ||
| 43 | RAB3IL1 | 3178 | 2.046 | 0.4458 | No | ||
| 44 | SYT6 | 3751 | 1.680 | 0.4182 | No | ||
| 45 | FAF1 | 3909 | 1.598 | 0.4129 | No | ||
| 46 | RFX5 | 3954 | 1.579 | 0.4137 | No | ||
| 47 | WEE1 | 3977 | 1.567 | 0.4156 | No | ||
| 48 | LMX1A | 4025 | 1.546 | 0.4162 | No | ||
| 49 | RORC | 4040 | 1.540 | 0.4185 | No | ||
| 50 | PPP1R3C | 4209 | 1.468 | 0.4123 | No | ||
| 51 | B3GALT2 | 4385 | 1.397 | 0.4057 | No | ||
| 52 | CFL1 | 4411 | 1.389 | 0.4071 | No | ||
| 53 | HOXC13 | 4487 | 1.355 | 0.4058 | No | ||
| 54 | STMN1 | 4636 | 1.302 | 0.4004 | No | ||
| 55 | AVPI1 | 4714 | 1.270 | 0.3987 | No | ||
| 56 | UBE4B | 4790 | 1.238 | 0.3972 | No | ||
| 57 | CTBP2 | 4805 | 1.231 | 0.3989 | No | ||
| 58 | CUL5 | 4870 | 1.206 | 0.3978 | No | ||
| 59 | GPD1 | 4889 | 1.201 | 0.3993 | No | ||
| 60 | BDNF | 4945 | 1.182 | 0.3987 | No | ||
| 61 | COPZ1 | 5200 | 1.107 | 0.3871 | No | ||
| 62 | TIAL1 | 5227 | 1.100 | 0.3879 | No | ||
| 63 | LHX9 | 5282 | 1.086 | 0.3872 | No | ||
| 64 | CPT1A | 5691 | 0.956 | 0.3670 | No | ||
| 65 | SC5DL | 5779 | 0.932 | 0.3641 | No | ||
| 66 | CNNM1 | 5886 | 0.900 | 0.3602 | No | ||
| 67 | SLCO1C1 | 5970 | 0.877 | 0.3575 | No | ||
| 68 | IPO7 | 6015 | 0.863 | 0.3568 | No | ||
| 69 | FCHSD2 | 6274 | 0.793 | 0.3444 | No | ||
| 70 | LZTS2 | 6382 | 0.769 | 0.3402 | No | ||
| 71 | RASGRP2 | 6556 | 0.723 | 0.3322 | No | ||
| 72 | FXYD6 | 6750 | 0.675 | 0.3231 | No | ||
| 73 | SLC16A1 | 6767 | 0.668 | 0.3236 | No | ||
| 74 | RTN4RL2 | 6890 | 0.635 | 0.3183 | No | ||
| 75 | PITX3 | 6988 | 0.609 | 0.3142 | No | ||
| 76 | HNRPF | 7168 | 0.567 | 0.3057 | No | ||
| 77 | PLA2G4A | 7230 | 0.552 | 0.3035 | No | ||
| 78 | HOXC12 | 7480 | 0.488 | 0.2910 | No | ||
| 79 | GUCY1A2 | 7573 | 0.468 | 0.2869 | No | ||
| 80 | FADS3 | 7880 | 0.386 | 0.2711 | No | ||
| 81 | KRTCAP2 | 7889 | 0.384 | 0.2715 | No | ||
| 82 | LTBR | 7993 | 0.359 | 0.2666 | No | ||
| 83 | ADAM12 | 8127 | 0.326 | 0.2600 | No | ||
| 84 | TGFB2 | 8377 | 0.261 | 0.2471 | No | ||
| 85 | POGK | 8472 | 0.240 | 0.2425 | No | ||
| 86 | FOSL1 | 9050 | 0.097 | 0.2114 | No | ||
| 87 | POU3F1 | 9135 | 0.075 | 0.2070 | No | ||
| 88 | ATP5F1 | 9182 | 0.061 | 0.2046 | No | ||
| 89 | TESK2 | 9279 | 0.037 | 0.1995 | No | ||
| 90 | LRFN4 | 9370 | 0.014 | 0.1946 | No | ||
| 91 | RARG | 9458 | -0.011 | 0.1900 | No | ||
| 92 | IPO13 | 9493 | -0.020 | 0.1882 | No | ||
| 93 | RPL22 | 9540 | -0.034 | 0.1857 | No | ||
| 94 | NRAS | 9596 | -0.046 | 0.1828 | No | ||
| 95 | HPCA | 9840 | -0.111 | 0.1699 | No | ||
| 96 | OPRD1 | 10062 | -0.168 | 0.1583 | No | ||
| 97 | UBXD3 | 10250 | -0.211 | 0.1485 | No | ||
| 98 | PTPRF | 10334 | -0.228 | 0.1445 | No | ||
| 99 | ARHGAP12 | 10444 | -0.249 | 0.1391 | No | ||
| 100 | PAX6 | 10602 | -0.286 | 0.1312 | No | ||
| 101 | NRIP3 | 10659 | -0.297 | 0.1287 | No | ||
| 102 | USP2 | 10680 | -0.302 | 0.1283 | No | ||
| 103 | RBBP4 | 10886 | -0.354 | 0.1179 | No | ||
| 104 | HTATIP | 10913 | -0.364 | 0.1172 | No | ||
| 105 | AGMAT | 11205 | -0.436 | 0.1023 | No | ||
| 106 | NKX2-3 | 11279 | -0.454 | 0.0992 | No | ||
| 107 | HMGN2 | 11287 | -0.455 | 0.0998 | No | ||
| 108 | SLC1A7 | 11292 | -0.456 | 0.1005 | No | ||
| 109 | CGN | 11457 | -0.498 | 0.0926 | No | ||
| 110 | LIN28 | 11552 | -0.520 | 0.0885 | No | ||
| 111 | TEX12 | 11804 | -0.593 | 0.0761 | No | ||
| 112 | ATF1 | 11838 | -0.603 | 0.0755 | No | ||
| 113 | COL2A1 | 11924 | -0.626 | 0.0722 | No | ||
| 114 | ATP6V0B | 12166 | -0.694 | 0.0605 | No | ||
| 115 | CTSF | 12201 | -0.703 | 0.0601 | No | ||
| 116 | BAI2 | 12535 | -0.810 | 0.0437 | No | ||
| 117 | BRDT | 12910 | -0.918 | 0.0253 | No | ||
| 118 | NET1 | 12970 | -0.934 | 0.0239 | No | ||
| 119 | TAF6L | 13014 | -0.948 | 0.0235 | No | ||
| 120 | ALX4 | 13016 | -0.949 | 0.0254 | No | ||
| 121 | SCYL1 | 13169 | -1.005 | 0.0191 | No | ||
| 122 | NR0B2 | 13207 | -1.018 | 0.0192 | No | ||
| 123 | SEC23IP | 13297 | -1.051 | 0.0165 | No | ||
| 124 | B4GALT2 | 13365 | -1.074 | 0.0150 | No | ||
| 125 | APOA5 | 13574 | -1.159 | 0.0060 | No | ||
| 126 | TDRD1 | 13791 | -1.247 | -0.0032 | No | ||
| 127 | FOXD3 | 13823 | -1.256 | -0.0023 | No | ||
| 128 | CHRM1 | 13857 | -1.270 | -0.0016 | No | ||
| 129 | TRIM3 | 13999 | -1.321 | -0.0065 | No | ||
| 130 | LRP8 | 14231 | -1.409 | -0.0162 | No | ||
| 131 | SLC38A2 | 14261 | -1.421 | -0.0149 | No | ||
| 132 | FXYD2 | 14333 | -1.452 | -0.0159 | No | ||
| 133 | CBARA1 | 14356 | -1.461 | -0.0141 | No | ||
| 134 | RUSC1 | 14473 | -1.511 | -0.0174 | No | ||
| 135 | AP3M1 | 14530 | -1.538 | -0.0173 | No | ||
| 136 | TMEM24 | 14681 | -1.620 | -0.0222 | No | ||
| 137 | HPCAL4 | 14722 | -1.637 | -0.0211 | No | ||
| 138 | CBX5 | 15290 | -2.008 | -0.0478 | No | ||
| 139 | STX6 | 15414 | -2.097 | -0.0502 | No | ||
| 140 | TRIM46 | 15852 | -2.479 | -0.0689 | No | ||
| 141 | HOXC5 | 15939 | -2.582 | -0.0684 | No | ||
| 142 | CDC14A | 16035 | -2.699 | -0.0681 | No | ||
| 143 | ILK | 16089 | -2.770 | -0.0655 | No | ||
| 144 | NIT1 | 16219 | -2.927 | -0.0666 | No | ||
| 145 | SCAMP3 | 16230 | -2.943 | -0.0612 | No | ||
| 146 | PFKFB3 | 16375 | -3.169 | -0.0626 | No | ||
| 147 | BCL9L | 16456 | -3.299 | -0.0604 | No | ||
| 148 | BLOC1S1 | 16512 | -3.400 | -0.0565 | No | ||
| 149 | BCL9 | 16577 | -3.526 | -0.0529 | No | ||
| 150 | COMMD3 | 16590 | -3.544 | -0.0464 | No | ||
| 151 | RAD9A | 16717 | -3.761 | -0.0457 | No | ||
| 152 | SIRT1 | 16844 | -4.003 | -0.0445 | No | ||
| 153 | H3F3A | 17152 | -4.820 | -0.0515 | No | ||
| 154 | HLX1 | 17242 | -5.043 | -0.0462 | No | ||
| 155 | SORL1 | 17378 | -5.468 | -0.0425 | No | ||
| 156 | RRAGC | 17384 | -5.489 | -0.0317 | No | ||
| 157 | ESRRA | 17475 | -5.806 | -0.0250 | No | ||
| 158 | ARPC5 | 17697 | -6.518 | -0.0238 | No | ||
| 159 | HHEX | 17732 | -6.662 | -0.0123 | No | ||
| 160 | PSEN2 | 17754 | -6.742 | 0.0001 | No | ||
| 161 | ATF7IP | 17898 | -7.456 | 0.0073 | No | ||
| 162 | EPC1 | 17947 | -7.671 | 0.0201 | No | ||
| 163 | CDKN2C | 18019 | -8.014 | 0.0324 | No |