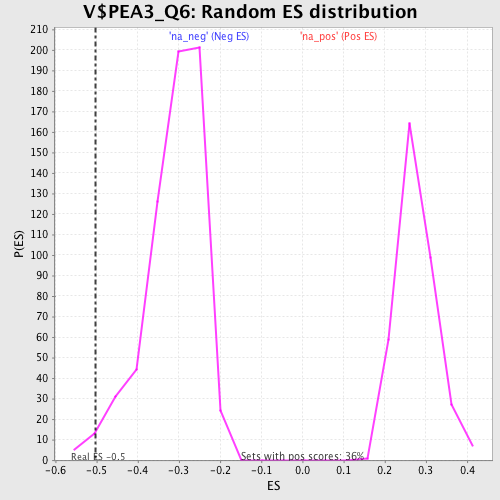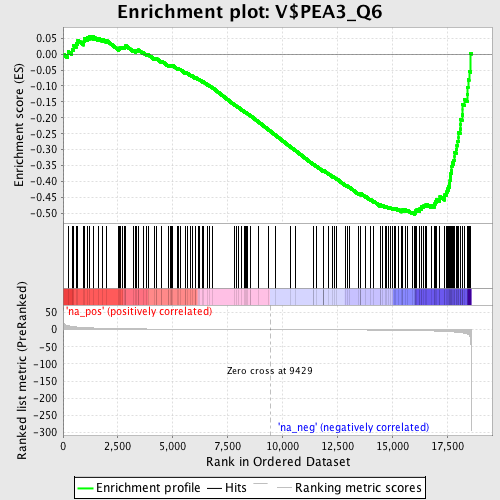
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$PEA3_Q6 |
| Enrichment Score (ES) | -0.5033541 |
| Normalized Enrichment Score (NES) | -1.6172798 |
| Nominal p-value | 0.01399689 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SELL | 222 | 11.135 | 0.0083 | No | ||
| 2 | KCNAB2 | 405 | 9.127 | 0.0151 | No | ||
| 3 | EXTL2 | 475 | 8.504 | 0.0269 | No | ||
| 4 | PICALM | 593 | 7.642 | 0.0345 | No | ||
| 5 | GTF2H1 | 671 | 7.198 | 0.0435 | No | ||
| 6 | CENTD2 | 950 | 5.981 | 0.0393 | No | ||
| 7 | CUGBP1 | 969 | 5.911 | 0.0491 | No | ||
| 8 | FEN1 | 1108 | 5.410 | 0.0515 | No | ||
| 9 | PRDX6 | 1193 | 5.127 | 0.0563 | No | ||
| 10 | ABLIM1 | 1381 | 4.578 | 0.0546 | No | ||
| 11 | SLC30A7 | 1605 | 4.048 | 0.0499 | No | ||
| 12 | TNNI2 | 1808 | 3.642 | 0.0456 | No | ||
| 13 | DGKZ | 1970 | 3.347 | 0.0430 | No | ||
| 14 | ANKRD1 | 2541 | 2.579 | 0.0168 | No | ||
| 15 | PAFAH2 | 2583 | 2.537 | 0.0192 | No | ||
| 16 | IL18BP | 2637 | 2.490 | 0.0209 | No | ||
| 17 | TYSND1 | 2700 | 2.441 | 0.0220 | No | ||
| 18 | SLC35C1 | 2812 | 2.336 | 0.0202 | No | ||
| 19 | DPP3 | 2825 | 2.329 | 0.0238 | No | ||
| 20 | SLC2A3 | 2859 | 2.294 | 0.0262 | No | ||
| 21 | TAF5 | 3186 | 2.040 | 0.0123 | No | ||
| 22 | GFI1 | 3306 | 1.943 | 0.0094 | No | ||
| 23 | LAG3 | 3351 | 1.912 | 0.0105 | No | ||
| 24 | RUNX3 | 3422 | 1.865 | 0.0101 | No | ||
| 25 | RPS3 | 3431 | 1.855 | 0.0131 | No | ||
| 26 | ELAVL4 | 3646 | 1.739 | 0.0047 | No | ||
| 27 | NUMA1 | 3807 | 1.650 | -0.0010 | No | ||
| 28 | RIN1 | 3887 | 1.610 | -0.0023 | No | ||
| 29 | MFN2 | 4149 | 1.498 | -0.0138 | No | ||
| 30 | DMBT1 | 4238 | 1.456 | -0.0159 | No | ||
| 31 | TNFRSF19L | 4246 | 1.452 | -0.0136 | No | ||
| 32 | LAMC1 | 4463 | 1.371 | -0.0228 | No | ||
| 33 | MMP7 | 4506 | 1.350 | -0.0226 | No | ||
| 34 | CTBP2 | 4805 | 1.231 | -0.0365 | No | ||
| 35 | PLCB3 | 4807 | 1.229 | -0.0343 | No | ||
| 36 | GPD1 | 4889 | 1.201 | -0.0365 | No | ||
| 37 | RNASEL | 4937 | 1.185 | -0.0369 | No | ||
| 38 | BDNF | 4945 | 1.182 | -0.0351 | No | ||
| 39 | CD5 | 4987 | 1.170 | -0.0352 | No | ||
| 40 | TIAL1 | 5227 | 1.100 | -0.0461 | No | ||
| 41 | MAP4K2 | 5249 | 1.096 | -0.0453 | No | ||
| 42 | FGF23 | 5330 | 1.073 | -0.0476 | No | ||
| 43 | SLC26A9 | 5589 | 0.987 | -0.0598 | No | ||
| 44 | ANK3 | 5594 | 0.986 | -0.0582 | No | ||
| 45 | EHF | 5656 | 0.967 | -0.0598 | No | ||
| 46 | ADAMTS4 | 5809 | 0.923 | -0.0663 | No | ||
| 47 | FUCA1 | 5878 | 0.902 | -0.0684 | No | ||
| 48 | IPO7 | 6015 | 0.863 | -0.0742 | No | ||
| 49 | CAPZA1 | 6044 | 0.856 | -0.0741 | No | ||
| 50 | GPA33 | 6158 | 0.823 | -0.0787 | No | ||
| 51 | CPNE8 | 6205 | 0.813 | -0.0797 | No | ||
| 52 | SNCG | 6368 | 0.771 | -0.0871 | No | ||
| 53 | LZTS2 | 6382 | 0.769 | -0.0864 | No | ||
| 54 | VCAM1 | 6600 | 0.710 | -0.0969 | No | ||
| 55 | CYR61 | 6688 | 0.687 | -0.1003 | No | ||
| 56 | NCAM1 | 6798 | 0.659 | -0.1050 | No | ||
| 57 | GPR120 | 7788 | 0.406 | -0.1579 | No | ||
| 58 | PGM2L1 | 7904 | 0.380 | -0.1634 | No | ||
| 59 | LTBR | 7993 | 0.359 | -0.1675 | No | ||
| 60 | SV2A | 8152 | 0.317 | -0.1755 | No | ||
| 61 | FCGR3A | 8278 | 0.282 | -0.1818 | No | ||
| 62 | RPS6KA4 | 8330 | 0.271 | -0.1840 | No | ||
| 63 | PRF1 | 8338 | 0.269 | -0.1839 | No | ||
| 64 | VDAC2 | 8383 | 0.260 | -0.1858 | No | ||
| 65 | CD3E | 8523 | 0.227 | -0.1929 | No | ||
| 66 | CTSW | 8532 | 0.223 | -0.1930 | No | ||
| 67 | XCL1 | 8910 | 0.136 | -0.2131 | No | ||
| 68 | LRFN4 | 9370 | 0.014 | -0.2380 | No | ||
| 69 | RAB39 | 9680 | -0.067 | -0.2546 | No | ||
| 70 | CBL | 10381 | -0.236 | -0.2921 | No | ||
| 71 | PAX6 | 10602 | -0.286 | -0.3035 | No | ||
| 72 | MMRN2 | 11403 | -0.484 | -0.3460 | No | ||
| 73 | KCNQ1 | 11424 | -0.489 | -0.3461 | No | ||
| 74 | LIN28 | 11552 | -0.520 | -0.3521 | No | ||
| 75 | SYTL1 | 11849 | -0.605 | -0.3670 | No | ||
| 76 | HIPK1 | 11861 | -0.608 | -0.3665 | No | ||
| 77 | GATA3 | 11864 | -0.608 | -0.3655 | No | ||
| 78 | SRRM1 | 11871 | -0.610 | -0.3647 | No | ||
| 79 | MARK1 | 12096 | -0.675 | -0.3756 | No | ||
| 80 | SLC41A1 | 12289 | -0.728 | -0.3847 | No | ||
| 81 | SPATA6 | 12348 | -0.750 | -0.3865 | No | ||
| 82 | GRIN2B | 12460 | -0.783 | -0.3910 | No | ||
| 83 | TRIM33 | 12860 | -0.905 | -0.4110 | No | ||
| 84 | GAB2 | 12943 | -0.928 | -0.4137 | No | ||
| 85 | IVL | 13040 | -0.954 | -0.4172 | No | ||
| 86 | CACNA1E | 13481 | -1.118 | -0.4390 | No | ||
| 87 | LALBA | 13544 | -1.147 | -0.4403 | No | ||
| 88 | BCL2L14 | 13553 | -1.151 | -0.4386 | No | ||
| 89 | NDUFS2 | 13555 | -1.152 | -0.4366 | No | ||
| 90 | INPPL1 | 13790 | -1.247 | -0.4470 | No | ||
| 91 | SPRR4 | 14027 | -1.333 | -0.4573 | No | ||
| 92 | SELP | 14164 | -1.385 | -0.4622 | No | ||
| 93 | ARHGDIB | 14456 | -1.505 | -0.4752 | No | ||
| 94 | PUM1 | 14466 | -1.509 | -0.4729 | No | ||
| 95 | CTSS | 14578 | -1.555 | -0.4761 | No | ||
| 96 | TMEM24 | 14681 | -1.620 | -0.4786 | No | ||
| 97 | ARF3 | 14752 | -1.654 | -0.4794 | No | ||
| 98 | SEC24C | 14844 | -1.710 | -0.4812 | No | ||
| 99 | ELK4 | 14933 | -1.758 | -0.4828 | No | ||
| 100 | LPXN | 15017 | -1.808 | -0.4840 | No | ||
| 101 | LRMP | 15096 | -1.859 | -0.4848 | No | ||
| 102 | PBXIP1 | 15141 | -1.893 | -0.4837 | No | ||
| 103 | PDGFD | 15301 | -2.013 | -0.4887 | No | ||
| 104 | KAZALD1 | 15429 | -2.109 | -0.4917 | No | ||
| 105 | ADRBK1 | 15471 | -2.148 | -0.4900 | No | ||
| 106 | MR1 | 15484 | -2.155 | -0.4867 | No | ||
| 107 | SIPA1 | 15599 | -2.247 | -0.4888 | No | ||
| 108 | ETV3 | 15693 | -2.337 | -0.4896 | No | ||
| 109 | MARK2 | 15906 | -2.543 | -0.4964 | No | ||
| 110 | CDC14A | 16035 | -2.699 | -0.4984 | Yes | ||
| 111 | ITPR2 | 16048 | -2.720 | -0.4941 | Yes | ||
| 112 | TCIRG1 | 16075 | -2.748 | -0.4905 | Yes | ||
| 113 | VPS11 | 16125 | -2.809 | -0.4880 | Yes | ||
| 114 | XPNPEP1 | 16254 | -2.970 | -0.4895 | Yes | ||
| 115 | FUT11 | 16262 | -2.983 | -0.4845 | Yes | ||
| 116 | ITPKB | 16338 | -3.119 | -0.4828 | Yes | ||
| 117 | TRIM8 | 16347 | -3.129 | -0.4776 | Yes | ||
| 118 | PTPRC | 16439 | -3.278 | -0.4765 | Yes | ||
| 119 | ELOVL1 | 16497 | -3.373 | -0.4734 | Yes | ||
| 120 | ZRANB1 | 16581 | -3.533 | -0.4715 | Yes | ||
| 121 | CD84 | 16779 | -3.884 | -0.4751 | Yes | ||
| 122 | PRKACB | 16911 | -4.157 | -0.4746 | Yes | ||
| 123 | POU2AF1 | 16943 | -4.236 | -0.4685 | Yes | ||
| 124 | LCK | 16984 | -4.337 | -0.4628 | Yes | ||
| 125 | BLNK | 17018 | -4.436 | -0.4565 | Yes | ||
| 126 | IRAK4 | 17160 | -4.845 | -0.4553 | Yes | ||
| 127 | UNC93B1 | 17174 | -4.875 | -0.4471 | Yes | ||
| 128 | SORL1 | 17378 | -5.468 | -0.4481 | Yes | ||
| 129 | RHOG | 17401 | -5.529 | -0.4392 | Yes | ||
| 130 | ESRRA | 17475 | -5.806 | -0.4325 | Yes | ||
| 131 | PIK4CB | 17502 | -5.917 | -0.4231 | Yes | ||
| 132 | ST7L | 17577 | -6.163 | -0.4159 | Yes | ||
| 133 | CD69 | 17611 | -6.271 | -0.4062 | Yes | ||
| 134 | IL2RA | 17631 | -6.321 | -0.3957 | Yes | ||
| 135 | DCLRE1C | 17651 | -6.380 | -0.3851 | Yes | ||
| 136 | ZNF289 | 17672 | -6.427 | -0.3745 | Yes | ||
| 137 | CMAS | 17700 | -6.524 | -0.3640 | Yes | ||
| 138 | CAMK1D | 17714 | -6.598 | -0.3527 | Yes | ||
| 139 | HHEX | 17732 | -6.662 | -0.3414 | Yes | ||
| 140 | NT5C2 | 17807 | -6.965 | -0.3327 | Yes | ||
| 141 | ZDHHC5 | 17829 | -7.088 | -0.3209 | Yes | ||
| 142 | SPI1 | 17833 | -7.094 | -0.3081 | Yes | ||
| 143 | TCEB3 | 17917 | -7.568 | -0.2988 | Yes | ||
| 144 | EPC1 | 17947 | -7.671 | -0.2864 | Yes | ||
| 145 | CD53 | 17986 | -7.827 | -0.2742 | Yes | ||
| 146 | MAPKAPK2 | 18015 | -7.995 | -0.2611 | Yes | ||
| 147 | CDKN2C | 18019 | -8.014 | -0.2466 | Yes | ||
| 148 | SSBP3 | 18095 | -8.603 | -0.2350 | Yes | ||
| 149 | AKT3 | 18107 | -8.687 | -0.2197 | Yes | ||
| 150 | NCF2 | 18115 | -8.749 | -0.2041 | Yes | ||
| 151 | FLI1 | 18187 | -9.387 | -0.1908 | Yes | ||
| 152 | POLD4 | 18204 | -9.516 | -0.1743 | Yes | ||
| 153 | SLAMF9 | 18205 | -9.517 | -0.1570 | Yes | ||
| 154 | EPHA2 | 18292 | -10.437 | -0.1426 | Yes | ||
| 155 | WASF2 | 18418 | -12.638 | -0.1263 | Yes | ||
| 156 | PTPN6 | 18441 | -13.086 | -0.1036 | Yes | ||
| 157 | CAP1 | 18458 | -13.630 | -0.0796 | Yes | ||
| 158 | NRGN | 18502 | -15.718 | -0.0532 | Yes | ||
| 159 | IL18 | 18578 | -32.489 | 0.0021 | Yes |