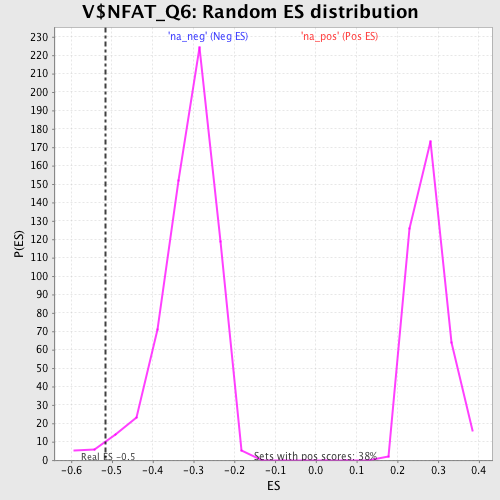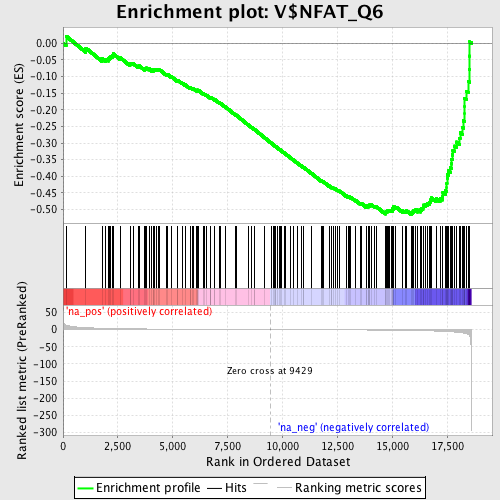
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFAT_Q6 |
| Enrichment Score (ES) | -0.51504636 |
| Normalized Enrichment Score (NES) | -1.6421978 |
| Nominal p-value | 0.019386107 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TNFAIP1 | 137 | 12.965 | 0.0215 | No | ||
| 2 | CDC2L2 | 1035 | 5.684 | -0.0143 | No | ||
| 3 | HSPG2 | 1776 | 3.719 | -0.0461 | No | ||
| 4 | HSD3B7 | 1943 | 3.391 | -0.0476 | No | ||
| 5 | ANKRD17 | 2053 | 3.218 | -0.0463 | No | ||
| 6 | GATA4 | 2122 | 3.119 | -0.0430 | No | ||
| 7 | SFRS1 | 2175 | 3.032 | -0.0390 | No | ||
| 8 | FOXP3 | 2247 | 2.919 | -0.0363 | No | ||
| 9 | AGTR2 | 2278 | 2.890 | -0.0315 | No | ||
| 10 | INPP5E | 2592 | 2.526 | -0.0428 | No | ||
| 11 | IL4 | 3049 | 2.137 | -0.0628 | No | ||
| 12 | EDN1 | 3081 | 2.118 | -0.0597 | No | ||
| 13 | MTIF2 | 3185 | 2.041 | -0.0607 | No | ||
| 14 | PAX3 | 3429 | 1.858 | -0.0697 | No | ||
| 15 | TACC2 | 3468 | 1.839 | -0.0677 | No | ||
| 16 | SLC26A7 | 3712 | 1.702 | -0.0771 | No | ||
| 17 | FRS3 | 3768 | 1.671 | -0.0763 | No | ||
| 18 | PER2 | 3798 | 1.654 | -0.0742 | No | ||
| 19 | NDRG2 | 3914 | 1.596 | -0.0768 | No | ||
| 20 | TBX4 | 4047 | 1.535 | -0.0806 | No | ||
| 21 | JDP2 | 4107 | 1.514 | -0.0804 | No | ||
| 22 | HOXC4 | 4118 | 1.511 | -0.0775 | No | ||
| 23 | HOXB3 | 4180 | 1.482 | -0.0775 | No | ||
| 24 | TRIM47 | 4278 | 1.437 | -0.0796 | No | ||
| 25 | ELF5 | 4329 | 1.418 | -0.0791 | No | ||
| 26 | NR3C2 | 4403 | 1.391 | -0.0800 | No | ||
| 27 | BNC2 | 4711 | 1.271 | -0.0938 | No | ||
| 28 | TNFRSF1A | 4774 | 1.243 | -0.0943 | No | ||
| 29 | SGOL1 | 4922 | 1.188 | -0.0996 | No | ||
| 30 | SPINK5 | 5217 | 1.103 | -0.1131 | No | ||
| 31 | TIAL1 | 5227 | 1.100 | -0.1111 | No | ||
| 32 | ZIC1 | 5427 | 1.034 | -0.1196 | No | ||
| 33 | TLL1 | 5562 | 0.994 | -0.1247 | No | ||
| 34 | CABP2 | 5790 | 0.928 | -0.1349 | No | ||
| 35 | UXS1 | 5800 | 0.926 | -0.1333 | No | ||
| 36 | TRPM3 | 5915 | 0.893 | -0.1375 | No | ||
| 37 | CAPN6 | 5944 | 0.887 | -0.1370 | No | ||
| 38 | PITX2 | 6070 | 0.848 | -0.1419 | No | ||
| 39 | PAK1IP1 | 6107 | 0.838 | -0.1420 | No | ||
| 40 | MAST2 | 6113 | 0.836 | -0.1404 | No | ||
| 41 | PTGER1 | 6185 | 0.817 | -0.1424 | No | ||
| 42 | PHOX2B | 6390 | 0.767 | -0.1517 | No | ||
| 43 | PLAC1 | 6441 | 0.754 | -0.1528 | No | ||
| 44 | LGALS1 | 6535 | 0.727 | -0.1562 | No | ||
| 45 | PCDH10 | 6714 | 0.682 | -0.1643 | No | ||
| 46 | ARHGAP5 | 6737 | 0.677 | -0.1640 | No | ||
| 47 | DTX3 | 6739 | 0.677 | -0.1625 | No | ||
| 48 | PCDH12 | 6910 | 0.630 | -0.1703 | No | ||
| 49 | IFNB1 | 6917 | 0.628 | -0.1692 | No | ||
| 50 | FAP | 7115 | 0.580 | -0.1786 | No | ||
| 51 | TRPS1 | 7169 | 0.567 | -0.1802 | No | ||
| 52 | RBMS3 | 7387 | 0.517 | -0.1908 | No | ||
| 53 | SOX10 | 7874 | 0.387 | -0.2163 | No | ||
| 54 | HOXA2 | 7879 | 0.386 | -0.2156 | No | ||
| 55 | IGFBP3 | 8459 | 0.244 | -0.2465 | No | ||
| 56 | ZIC4 | 8600 | 0.208 | -0.2536 | No | ||
| 57 | GHR | 8702 | 0.182 | -0.2587 | No | ||
| 58 | FJX1 | 8721 | 0.178 | -0.2592 | No | ||
| 59 | TEAD1 | 9190 | 0.059 | -0.2845 | No | ||
| 60 | CRY1 | 9475 | -0.015 | -0.2998 | No | ||
| 61 | ERG | 9592 | -0.046 | -0.3060 | No | ||
| 62 | PCBP2 | 9615 | -0.051 | -0.3071 | No | ||
| 63 | DEPDC2 | 9668 | -0.064 | -0.3098 | No | ||
| 64 | SLC6A12 | 9677 | -0.066 | -0.3100 | No | ||
| 65 | PLOD2 | 9790 | -0.096 | -0.3159 | No | ||
| 66 | TEK | 9850 | -0.115 | -0.3188 | No | ||
| 67 | NRK | 9887 | -0.124 | -0.3205 | No | ||
| 68 | SNCAIP | 9936 | -0.136 | -0.3228 | No | ||
| 69 | ADAMTS17 | 10074 | -0.172 | -0.3298 | No | ||
| 70 | PAK7 | 10147 | -0.187 | -0.3333 | No | ||
| 71 | SPARCL1 | 10359 | -0.233 | -0.3442 | No | ||
| 72 | DLL4 | 10514 | -0.263 | -0.3520 | No | ||
| 73 | USP2 | 10680 | -0.302 | -0.3603 | No | ||
| 74 | MAGED1 | 10843 | -0.343 | -0.3683 | No | ||
| 75 | MRGPRF | 10957 | -0.375 | -0.3736 | No | ||
| 76 | SOX3 | 10960 | -0.375 | -0.3728 | No | ||
| 77 | POU3F3 | 11335 | -0.469 | -0.3920 | No | ||
| 78 | WISP1 | 11754 | -0.577 | -0.4134 | No | ||
| 79 | SNX2 | 11816 | -0.595 | -0.4154 | No | ||
| 80 | GTF2E1 | 11854 | -0.606 | -0.4160 | No | ||
| 81 | CAPG | 12123 | -0.682 | -0.4290 | No | ||
| 82 | EXOSC9 | 12244 | -0.714 | -0.4339 | No | ||
| 83 | NFATC4 | 12252 | -0.716 | -0.4327 | No | ||
| 84 | RNF128 | 12334 | -0.746 | -0.4354 | No | ||
| 85 | MITF | 12395 | -0.766 | -0.4370 | No | ||
| 86 | ESRRB | 12527 | -0.809 | -0.4423 | No | ||
| 87 | DDR2 | 12598 | -0.829 | -0.4442 | No | ||
| 88 | SP3 | 12898 | -0.916 | -0.4584 | No | ||
| 89 | COL3A1 | 13023 | -0.950 | -0.4629 | No | ||
| 90 | SOX21 | 13030 | -0.952 | -0.4611 | No | ||
| 91 | MAP4K4 | 13118 | -0.983 | -0.4637 | No | ||
| 92 | CREB5 | 13324 | -1.060 | -0.4724 | No | ||
| 93 | CPNE1 | 13560 | -1.153 | -0.4826 | No | ||
| 94 | COL25A1 | 13621 | -1.181 | -0.4832 | No | ||
| 95 | GNAO1 | 13825 | -1.256 | -0.4914 | No | ||
| 96 | FOXP2 | 13833 | -1.260 | -0.4889 | No | ||
| 97 | TNXB | 13903 | -1.284 | -0.4898 | No | ||
| 98 | COL8A1 | 13920 | -1.292 | -0.4878 | No | ||
| 99 | PDGFB | 13980 | -1.314 | -0.4880 | No | ||
| 100 | JARID2 | 13982 | -1.315 | -0.4852 | No | ||
| 101 | CNTFR | 14034 | -1.336 | -0.4849 | No | ||
| 102 | FLT4 | 14183 | -1.394 | -0.4898 | No | ||
| 103 | P2RY1 | 14260 | -1.421 | -0.4908 | No | ||
| 104 | ACVR1 | 14685 | -1.621 | -0.5101 | Yes | ||
| 105 | BMP5 | 14704 | -1.629 | -0.5075 | Yes | ||
| 106 | POU4F2 | 14748 | -1.653 | -0.5061 | Yes | ||
| 107 | ARHGAP6 | 14779 | -1.669 | -0.5040 | Yes | ||
| 108 | PDE4D | 14829 | -1.703 | -0.5028 | Yes | ||
| 109 | IL2 | 14885 | -1.734 | -0.5019 | Yes | ||
| 110 | SULF1 | 14968 | -1.779 | -0.5024 | Yes | ||
| 111 | ANP32E | 15018 | -1.809 | -0.5010 | Yes | ||
| 112 | RBM14 | 15023 | -1.811 | -0.4972 | Yes | ||
| 113 | SHOX2 | 15067 | -1.840 | -0.4954 | Yes | ||
| 114 | ARF6 | 15073 | -1.843 | -0.4916 | Yes | ||
| 115 | COL16A1 | 15171 | -1.912 | -0.4925 | Yes | ||
| 116 | ADRBK1 | 15471 | -2.148 | -0.5039 | Yes | ||
| 117 | GPX1 | 15584 | -2.235 | -0.5050 | Yes | ||
| 118 | LIF | 15662 | -2.303 | -0.5040 | Yes | ||
| 119 | TEC | 15866 | -2.498 | -0.5095 | Yes | ||
| 120 | TGFB3 | 15935 | -2.580 | -0.5074 | Yes | ||
| 121 | FOXB1 | 15960 | -2.605 | -0.5029 | Yes | ||
| 122 | COMMD10 | 16042 | -2.711 | -0.5012 | Yes | ||
| 123 | LIMS1 | 16164 | -2.865 | -0.5013 | Yes | ||
| 124 | KIF3C | 16282 | -3.003 | -0.5010 | Yes | ||
| 125 | CFL2 | 16345 | -3.128 | -0.4973 | Yes | ||
| 126 | PHF6 | 16403 | -3.222 | -0.4932 | Yes | ||
| 127 | NR2F2 | 16413 | -3.231 | -0.4865 | Yes | ||
| 128 | HAPLN1 | 16516 | -3.409 | -0.4844 | Yes | ||
| 129 | PIK3CD | 16600 | -3.558 | -0.4810 | Yes | ||
| 130 | MRPL45 | 16702 | -3.738 | -0.4781 | Yes | ||
| 131 | PVRL1 | 16732 | -3.784 | -0.4712 | Yes | ||
| 132 | CD84 | 16779 | -3.884 | -0.4650 | Yes | ||
| 133 | FOXP1 | 17003 | -4.392 | -0.4673 | Yes | ||
| 134 | GADD45G | 17181 | -4.886 | -0.4659 | Yes | ||
| 135 | LENG4 | 17285 | -5.183 | -0.4599 | Yes | ||
| 136 | FBXL20 | 17308 | -5.257 | -0.4494 | Yes | ||
| 137 | GSK3B | 17428 | -5.589 | -0.4433 | Yes | ||
| 138 | VAMP8 | 17486 | -5.835 | -0.4334 | Yes | ||
| 139 | IRF1 | 17489 | -5.851 | -0.4204 | Yes | ||
| 140 | RAB6A | 17498 | -5.898 | -0.4077 | Yes | ||
| 141 | ZHX2 | 17508 | -5.940 | -0.3949 | Yes | ||
| 142 | INPP5F | 17555 | -6.081 | -0.3838 | Yes | ||
| 143 | HOXB4 | 17648 | -6.367 | -0.3746 | Yes | ||
| 144 | TIGD2 | 17699 | -6.524 | -0.3627 | Yes | ||
| 145 | TGFB1 | 17710 | -6.583 | -0.3485 | Yes | ||
| 146 | RHOA | 17735 | -6.687 | -0.3349 | Yes | ||
| 147 | LASP1 | 17764 | -6.772 | -0.3213 | Yes | ||
| 148 | KLF13 | 17831 | -7.092 | -0.3090 | Yes | ||
| 149 | BLR1 | 17922 | -7.579 | -0.2969 | Yes | ||
| 150 | TCTA | 18060 | -8.301 | -0.2858 | Yes | ||
| 151 | SSBP3 | 18095 | -8.603 | -0.2684 | Yes | ||
| 152 | ORMDL3 | 18200 | -9.480 | -0.2529 | Yes | ||
| 153 | ELK3 | 18247 | -9.988 | -0.2330 | Yes | ||
| 154 | EMP1 | 18289 | -10.408 | -0.2120 | Yes | ||
| 155 | EPHA2 | 18292 | -10.437 | -0.1888 | Yes | ||
| 156 | ZBTB5 | 18297 | -10.518 | -0.1655 | Yes | ||
| 157 | PTOV1 | 18382 | -11.849 | -0.1436 | Yes | ||
| 158 | IRF4 | 18485 | -15.042 | -0.1155 | Yes | ||
| 159 | MARCKS | 18519 | -17.134 | -0.0790 | Yes | ||
| 160 | SOX5 | 18531 | -18.503 | -0.0382 | Yes | ||
| 161 | ETV5 | 18537 | -19.144 | 0.0043 | Yes |